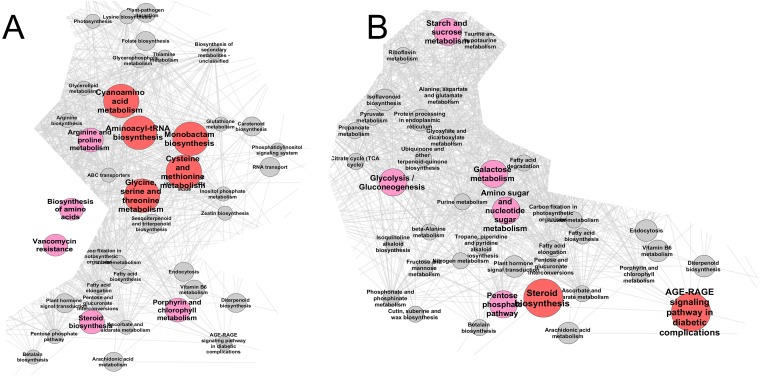
Figure 5. The effect of mycorrhiza development revealed by enrichment analysis (hypergeomertric test) on the metabolic pathways in the pea leaves of cv. Finale.
Fragment of pathway network based on the KEGG database using Medicago truncatula as a reference species. Nodes (pathways) share common edge if they share metabolites. The graph was built in the Cytoscape using Prefuse Layout, where lengths of edges reflect the number of metabolites shared between pathways. The larger red nodes correspond to p < 0.05, smaller pink nodes to p < 0.1, grey nodes to pathways sharing metabolites with significantly affected ones. (A) 42 DPI (stage IV); (B) 56 DPI (stage V). Full versions of the graphs are shown in the Fig. S6.