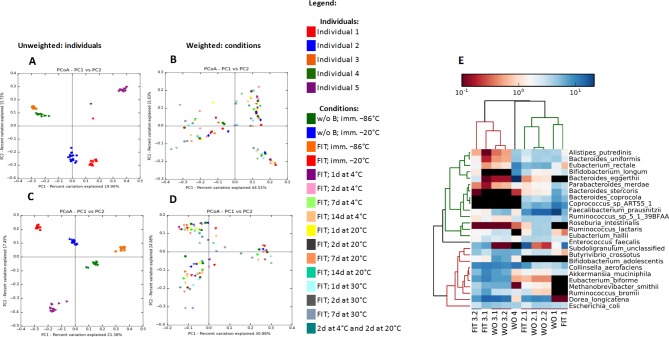
Figure 2.
Beta diversity (panels A–D) analysis of GI tract bacterial communities between the individuals and storage conditions presented in a form of principal coordinate analysis (PCoA) plot of weighted and unweighted UniFrac distances. Panels A and C are coloured according to the individual, panels B and D are coloured according to the conditions that samples were exposed to. Panels A and B represent the UniFrac metrics obtained from sequencing on the Illumina MiSeq platform, whereas panels C and D represent UniFrac metrics obtained from sequencing on the Ion Torrent PGM platform. The results of the metagenomics analysis at the species level are presented within panel E in a form of the heat-map profile for the most abundant entities in the metagenome samples. According to upper dendrogram, there is a consistency within the samples composition as the samples from the same individual are clustering together. Samples WO1, WO2.1, WO2.2, WO3.1, WO3.2, WO4 were without faecal immunochemical test (woFIT) and immediately frozen at −86°C; while samples FIT1, FIT2.1, FIT2.2, FIT3.1, FIT3.2 and FIT4 were with FIT (wFIT) and stored at 4°C for 2 days and then at 20°C for additional 2 days. Samples FIT2.2 and FIT4 failed at the sequencing stage and therefore are not presented within the figure. Both sample groups include technical replicates for individuals 2 and 3 (eg, WO2.1. and WO2.2.; WO3.1 and WO3.2.; FIT2.1 and FIT2.2.; FIT3.1. and FIT3.2.).