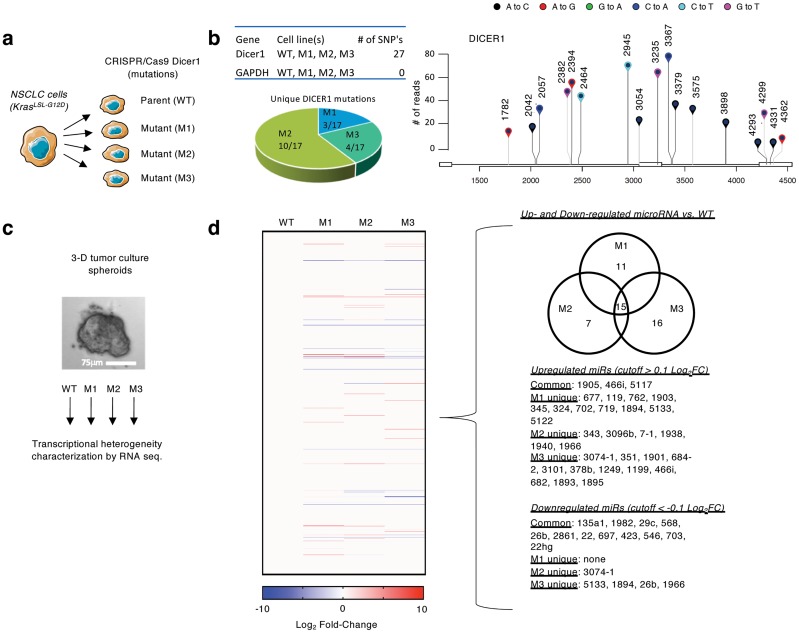
Fig 1. Characterizing phenotypic heterogeneity in Dicer1 engineered NSCLC cells.
a) Murine-derived non-small cell lung cancer (NSCLC) cells were engineered with mutant Dicer1 clones using the CRISPR/Cas9 toolkit. b) Lollipop graph quantifies SNPs in the mutant vs. WT cells including location and mutation annotation. Total number of normalized SNP counts is shown on the Y-axis. Table shows number of SNP identified in each cell line, GAPDH was used as a control to validate SNP identification algorithm. c) 3D cultured cells were characterized for transcriptional and therapeutic sensitivity heterogeneity. d) (left) Heat map shows differential expression pattern of miRNA compared to WT; (right) List of unique and shared differentially expressed miRNA.