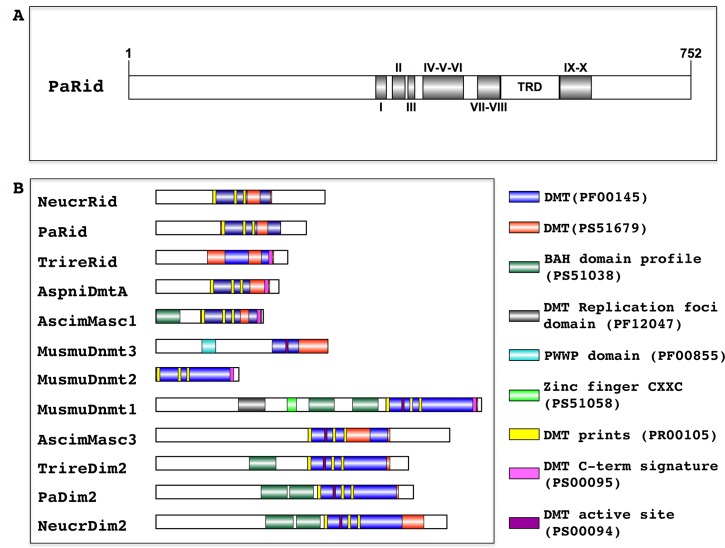
Fig 1. Structural analysis of DNA methyltransferase proteins.
(A) Domain architecture of P. anserina putative DNA methyltransferase PaRid. The catalytic domains contain 10 conserved motifs (I—X) and a target recognition domain (TRD) located between the motifs VIII and IX. The amino acid length is indicated. (B) Domain architecture of DNA methyltransferase proteins (DMT). The functional domain analysis was performed using InterProScan and visualized using IBS. Mus musculus: MusmuDnmt1: NP_001300940.1, MusmuDnmt2: NP_034197.3, MusmuDnmt3A: NP_031898.1; Ascobolus immersus, AscimMasc1 (AAC49849.1), AscimMasc3; Aspergillus nidulans AspniDmtA: XP_664242.1, Neurospora crassa NeucrRid: AAM27408.1, NeucrDim2: XP_959891.1; Podospora anserina PaDim2: Pa_5_9100; PaRid: Pa_1_19440; Trichoderma reesei TrireDim2 XP_006964860.1; TrireRid: AEM66210.1. Cytosine-specific DNA methyltransferase domains: PF00145, PR00105, PS51679, PS00095, PS00094; protein-DNA interaction domains: bromo-associated homology (BAH) domain PS51038, Replication foci targeting sequence (RFTS) PF12047, Zinc finger motif (CXXC) PS51058, PWWP domain (PF00855). Because its sequence is too divergent, PaDnmt5 (Pa_4_2960) was not included.