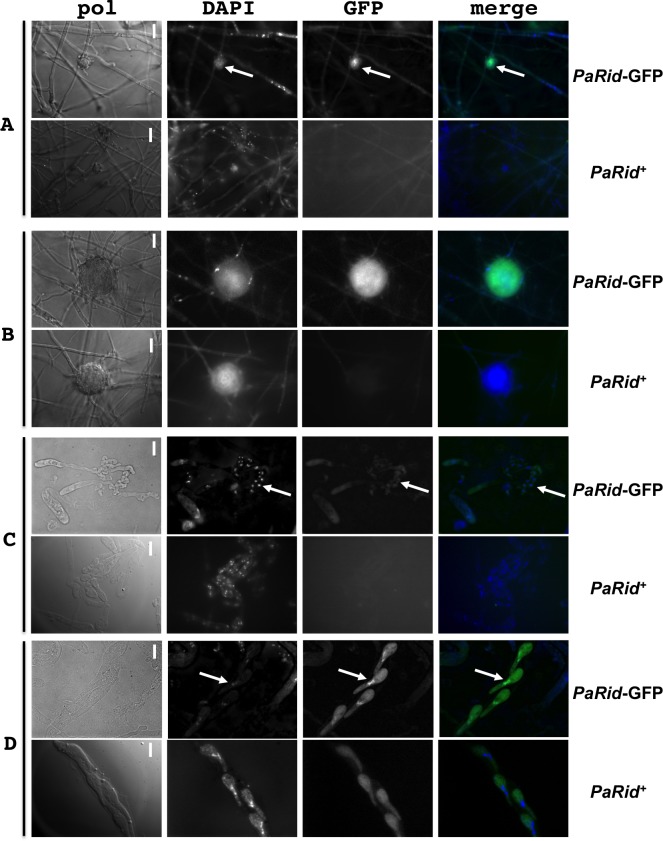
Fig 5. Expression and subcellular localization of PaRid during P. anserina life cycle.
PaRid-GFP expression was assayed from a ΔPaRid:AS4-PaRid-GFP-HA strain showing wild type phenotypes (PaRid-GFP). As a control, self-fluorescence was assayed from a wild-type strain (No GFP-tagged protein, PaRid+). No GFP signal can be observed in mycelium, however, a significant and specific GFP signal can be found in ascogonia (A, white arrows are pointing at ascogonia) and protoperithecia (B). Surprisingly, no GFP signal is observed in the croziers (C, white arrows are pointing at croziers) but a strong signal can be noticed in the mature ascospores (D, white arrows are pointing at the nuclei of one ascospore). Since self-fluorescence can be detected in the cytoplasm of ascospores, but not in the nuclei, the PaRid-GFP protein localization is nuclear. From left to right: bright-field (pol), DAPI staining, GFP channel and merge of the two latest. Scale bar: 1,5 μm (A and B), 5 μm (C and D).