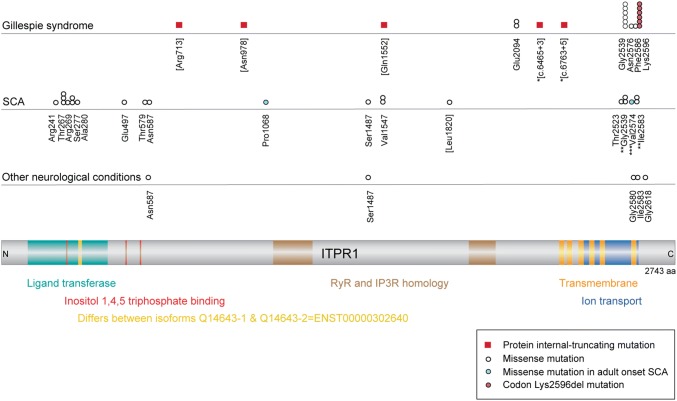
Fig. 5.
Spectrum of ITPR1 pathogenic variants. A linear protein schematic of ITPR1 is shown in the bottom panel, with domains and features demarcated and labelled in colour. The position of the 15-amino acid insertion in the longer isoform, UniProt Q14643-1, is shown in yellow. Amino acid numbering and domain positions are based on the 2743-amino acid isoform 2: Q14643-2, encoded by the canonical transcript GenBank NM_001168272.1; ENST00000302640. Shown above the protein schematic are all of the published variants associated with Gillespie syndrome and other neurological conditions, and all of the published substitution variants associated with spinocerebellar ataxia (SCA). All variants shown have had their amino acid numbering unified to isoform 2 (to facilitate an accurate collation and comparison of the ITPR1 dataset) and may therefore differ from the numbering used in the original publication. Variants in brackets indicate recessive alleles, of which *compound heterozygous alleles identified in a single proband; **cases reported as having a SCA29 phenotype and with substitutions at residues associated with Gillespie syndrome (identical variant) or pontocerebellar hypoplasia (different variant); ***variant associated with adult onset SCA in a mother and early onset SCA in her daughter