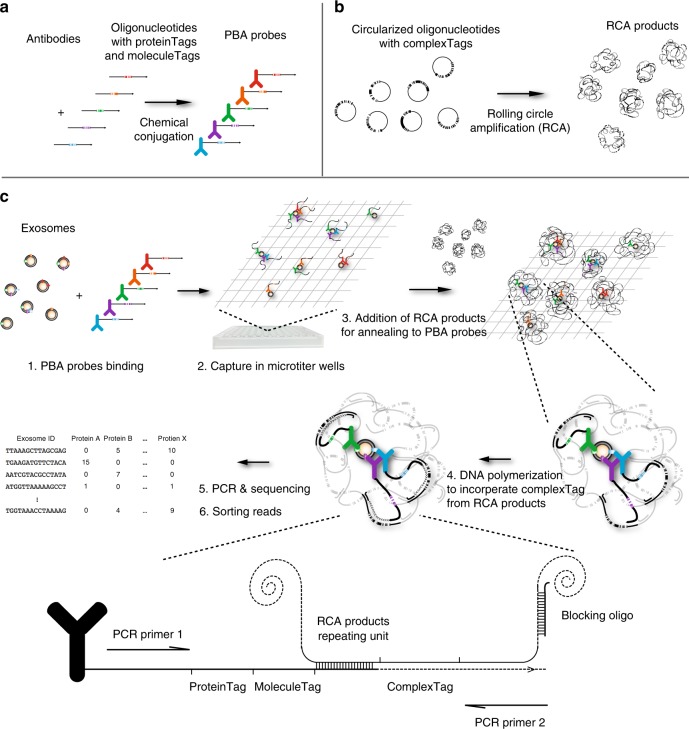
Fig. 1.
Design and workflow of PBA. a Preparation of PBA probes by chemical conjugation of antibodies and DNA oligonucleotides containing an 8-nt proteinTag and an 8-nt random sequence moleculeTag. b Preparation of RCA products from circularized oligonucleotides comprising a 15-nt random sequence as complexTags, capable of encoding in excess of one billion unique complexTags. c To profile surface proteins of exosomes by PBA, exosomes are first incubated with PBA probes, followed by capture of exosomes with bound PBA probes in microtiter wells via immobilized cholera toxin subunit B (CTB). Oligonucleotides on PBA probes brought together by binding the same exosome are next allowed to hybridize to a unique RCA product, followed by enzymatic extension, incorporating the complexTag present in the RCA product along with a standard sequence motif, later used for amplification. To prevent the DNA polymerase from extending across nearby monomers in the RCA products, blocking oligonucleotides are pre-hybridized to the RCA products. Successfully extended DNA molecules on PBA probes are amplified by PCR using the PCR primer1 and PCR primer2 for library preparation, while oligonucleotides on the antibodies or RCA products fail to be amplified by the PCR primer pairs used for amplification. The PCR product were subjected to DNA sequencing to record the numbers of molecules with specific tag combinations, thus revealing the identities of proteins on individual exosomes