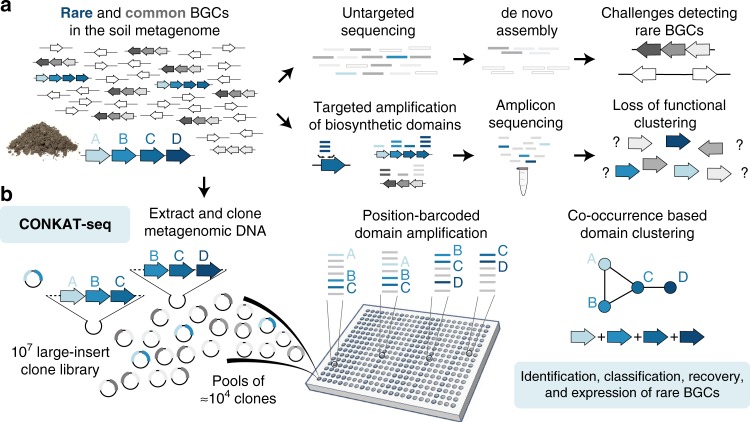
Fig. 1.
CONKAT-seq enables the exploration of rare biosynthetic gene clusters in complex metagenomes. a Untargeted methods to explore the biosynthetic potential of low frequency organisms in the soil metagenome are limited by the required high coverage depth, and computationally challenging de novo assembly process. PCR based methods are extremely sensitive, but do not capture the functional clustering of biosynthetic domain and therefore are information poor. b CONKAT-seq uses the highly partitioned structure of metagenomic cosmid libraries to reconstruct the chromosomal organization of biosynthetic domains based on PCR amplicon data