Fig. 3.
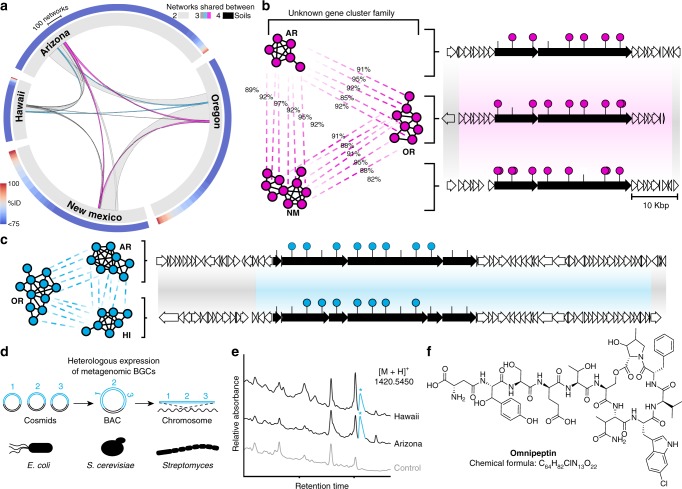
Comparing domain networks from different samples enables discovery of common yet unknown families of natural products. a 3591 domain networks obtained from four soil samples were compared to each other and to sequences of BGCs found in databases. For each soil sample, the outer ring (red and blue) depicts the proportions of networks that display similarity to sequences of BGC found in databases. The fraction of networks found in each soil sharing relatives with a median identity higher than 90% in other soil samples is represented by the ratio of the width of the ribbons relative to the corresponding part of the inner ring (gray). Each ribbon represents the fraction of networks shared by combinations of 2, 3, or all 4 soils analyzed. Colored ribbons represent networks with close relatives in Oregon, Arizona and New Mexico (Magenta) or Hawaii, Arizona and Oregon (Cyan) out of which one representative family was physically recovered and depicted in b and c. b Detailed view of the pairwise comparison of biosynthetic domains belonging to related networks that form a family of common yet unknown BGCs. Recovery of metagenomic clones encoding this BGC family from geographically distant soils revealed novel and highly similar gene content and architecture. Colored shading covers the portion of metagenomic DNA showing high homology between related BGCs while gray highlight genes lacking significant homology that likely belong to independent chromosomal contexts outside of BGC boundaries. c CONKAT-seq networks and BGCs for members of a family of common yet unknown BGCs identified in Hawaii and Arizona soil samples. d The corresponding overlapping cosmids isolated from E. coli clones were assembled by transformation-associated recombination in S. cerevisiae into a bacterial artificial chromosome (BAC) which was later integrated in the chromosome of Streptomyces albus. e Heterologous expression of the two related metagenomic BGCs and analysis of the associated crude extracts by HPLC/MS led to the detection of new peaks normally absent in the control extract obtained from the host native background (S. albus). f Both BGCs led to the isolation of the same major product, omnipeptin, a novel 11 residue cyclic depsipeptide