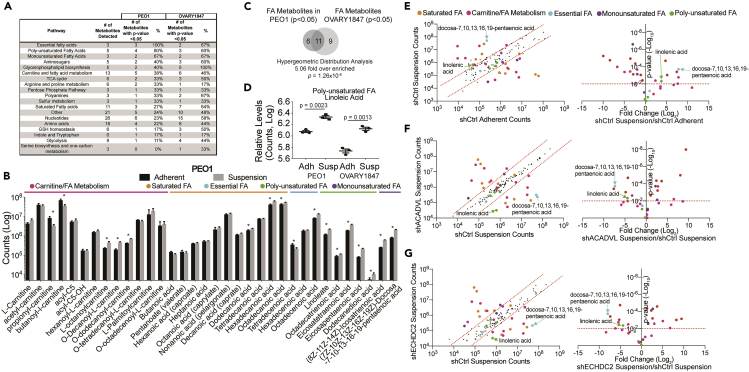
Figure 2.
HGSOC Cells Cultured in Suspension Are Enriched for Fatty Acids and Fatty Acid Metabolites
HGSOC cells (PEO1 and OVARY1847) were cultured in adherent and suspension cultures for 48 h. Cells were collected and used for global non-targeted metabolomics.
(A) Metabolites grouped based on pathway. The number of significant (p < 0.05) metabolites indicated with percentage (%) of pathway. See Table S1 for a full list of metabolites.
(B) Fatty acids and fatty acids metabolites in PEO1 cells grown in adherent (black bars) or suspension (gray bars) conditions. ∗p value < 0.05.
(C) Comparison of differentially enriched fatty acids and fatty acid metabolites in PEO1 versus OVARY1847.
(D) An example of poly-unsaturated fatty acids enriched in PEO1 and OVARY1847 suspension cells.
(E) Global non-targeted metabolomics analysis of shCtrl PEO1 cells cultured in adherent versus suspension. Left Panel, metabolite counts graphed as a scatterplot. x axis, shCtrl adherent and y axis, shCtrl suspension. Right Panel, x axis, Log2-Fold change of metabolites and y axis, p value.
(F) Metabolomics analysis of shCtrl PEO1 cells cultured in suspension versus shACADVL cells cultured in suspension. Left Panel, metabolite counts graphed as a scatterplot. x axis, shCtrl suspension and y axis, shACADVL suspension. Right Panel, x axis, Log2-Fold change of metabolites and y axis, p value.
(G) Metabolomics analysis of shCtrl PEO1 cells cultured in suspension versus shECHDC2 cells cultured in suspension. Left Panel, metabolite counts graphed as a scatterplot. x axis, shCtrl suspension and y axis, shECHDC2 suspension. Right Panel, x axis, Log2-Fold change of metabolites and y axis, p value. Red dotted lines indicated p value threshold. Statistical test = unpaired t test. Error bars = S.E.M.
See also Figure S2.