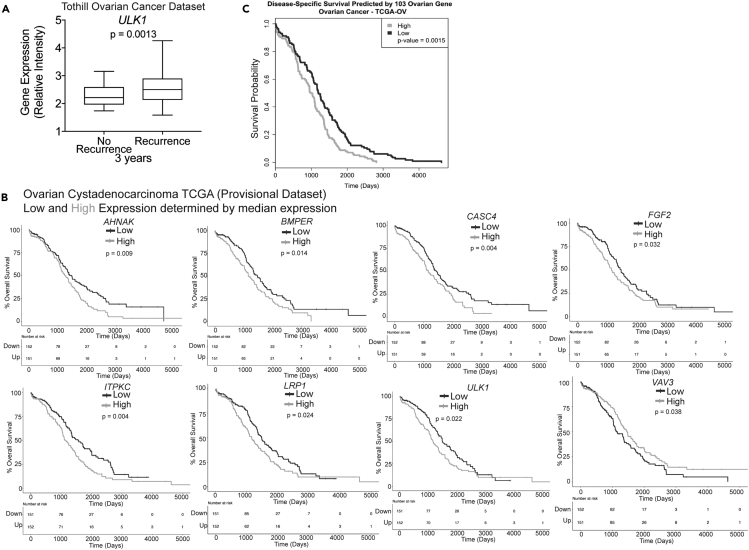
Figure 5.
ULK1 Predicts HGSOC Recurrence and Survival and the 108 Gene Signature Predicts Overall Survival
(A) Examination of ULK1 expression in primary ovarian adenocarcinoma that had no recurrence (n = 23) or recurrence (n = 96) after 3 years. Statistical test = two-sided unpaired t test. Error bars = S.E.M.
(B) A median expression was determined from each indicated gene to distinguish patient tumors with high expression (High, gray line) versus low expression (Low, dark line). KM curves were generated. TCGA (Ovarian cancer, Cancer Genome Atlas Research Network, 2011) dataset was used. Statistical test = Wilcoxon rank sum.
(C) For the TCGA Ovarian Cystadenocarcinoma dataset, 103 genes were mapped to their associated probe set or if the reference dataset was a list of genes, then expression data were pulled out based on matching gene names. Of the candidate genes, there was a varying number of probe sets present on all array platforms. Number of probe sets used indicated above KM curves. For each dataset, modeling survival and generating a KM curve on the candidate gene signature score. The dichotomous description of gene signature score (high or low gene signature score) was assigned. p Values were taken from the log likelihood statistic from the Cox proportional hazard models. For verification, we permuted a subset of random expression and random outcome (time, vital status) values, and broke the relationship between expression and outcome. This was done 1,000 times, and for each permuted dataset we modeled survival as described for the original analysis and generated a distribution of log likelihood statistics. Note: not all genes were identified in each dataset.