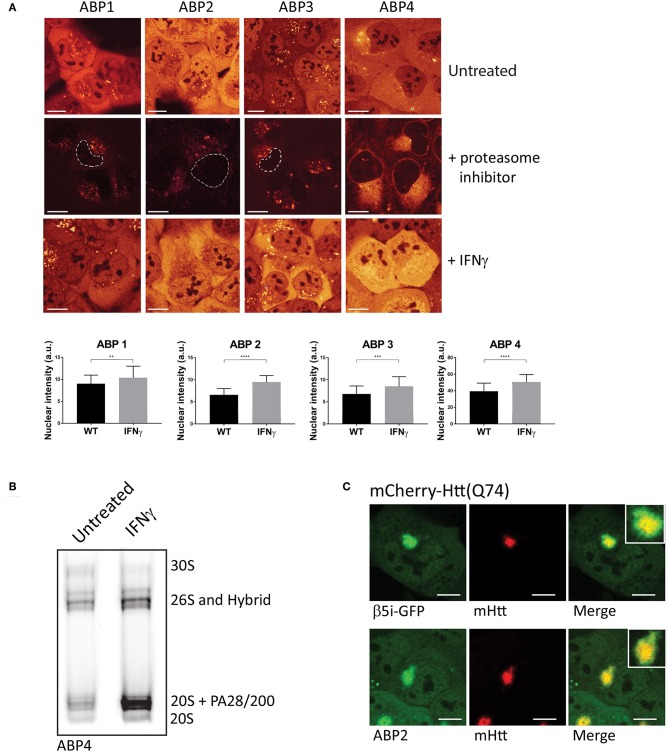
Figure 3.
Visualizing proteasome activity in living cells. (A) Proteasome activity labeling in living cells by different probes. U2OS cells were incubated with vinyl sulphone (ABP1, green and ABP3, red) and epoxomicin (ABP2, green and ABP4, yellow) based probes (upper panel). Pre-incubation with epoxomycin to block proteasome activity was used to determine nonspecific binding (middle panel). Epoxomycin-based probes give more intense labeling pattern, while the Cy3 fluorophore gives more background staining. When U2OS cells were stimulated for 72 h with IFN-γ, subsequent activity labeling showed a significant increase in labeling (lower panel, graphs). (B) U2OS cells were stimulated with IFN-γ, labeled with ABP4 and analyzed by native PAGE, confirming increased ABP labeling as shown by microscopy. (C) Recruitment of active proteasomes into aggregates. U2OS cells were transfected with polyglutamine-expanded huntingtin fragments to initiate aggregation, and co-transfected with β5i-GFP to show proteasome distribution around aggregates. Incubation with ABP2 showed a similar distribution pattern as β5i-GFP, indicating the recruitment of catalytically active proteasomes into aggregates. Scale bar = 5 μm.