Abstract
Cyclophosphamide is an alkylating agent used in the treatment of solid and haematological malignancies and as an immunosuppressive agent. As a prodrug, it is dependent on bioactivation to the active phosphoramide mustard metabolite to elicit its therapeutic effect. This focused review will highlight the evidence for the role of germline pharmacogenetic variation in both plasma pharmacokinetics and clinical outcomes. There is a substantial indication from 13 pharmacokinetic and 17 therapeutic outcome studies, in contexts as diverse as haematological malignancy, breast cancer, systemic lupus erythematosus and myeloablation, that pharmacogenetic variation in both CYP2C19 and CYP2B6 influence the bioactivation of cyclophosphamide. An additional role for pharmacogenetic variation in ALDH1A1 has also been reported. Future studies should comprehensively assess these 3 pharmacogenes and undertake appropriate statistical analysis of gene–gene interactions to confirm these findings and may allow personalised treatment regimens.
Keywords: chemotherapy, cytochrome P450 enzymes, genetic polymorphism, genetics and pharmacogenetics, oncology
1. INTRODUCTION
First synthesized over 60 years ago, cyclophosphamide (N,N‐bis(2‐chloroethyl)‐2‐oxo‐1,3,2λ5‐oxazaphosphinan‐2‐amine) remains a key chemotherapeutic agent for the treatment of a number of solid and haematological cancers inflammatory and autoimmune disorders, bone marrow transplantation (stem cell mobilization and conditioning regimens), and as prophylaxis against post‐transplantation graft‐vs‐host disease.1, 2, 3, 4, 5
Cyclophosphamide is a prodrug that requires bioactivation to elicit its therapeutic effects. The complex activation and inactivation pathways for this drug, and the enzymes or chemical reactions involved, are summarised in Figure 1. Cyclophosphamide undergoes hepatic hydroxylation to form 4‐hydroxycyclophosphamide (4‐OHCP), which represents the major route of metabolism of the drug and accounts for 70–80% of the dose.6 This route can be catalysed by any a number of cytochrome P450 (CYP) enzymes including CYP2B6, CYP2C9, CYP2C19, CYP3A4, CYP3A5 and CYP2J2.8, 9, 10, 11, 12, 13, 14, 15, 16 CYP2B6 and CYP2C19 are the enzymes with the highest activity for bioactivation of cyclophosphamide.
Figure 1.
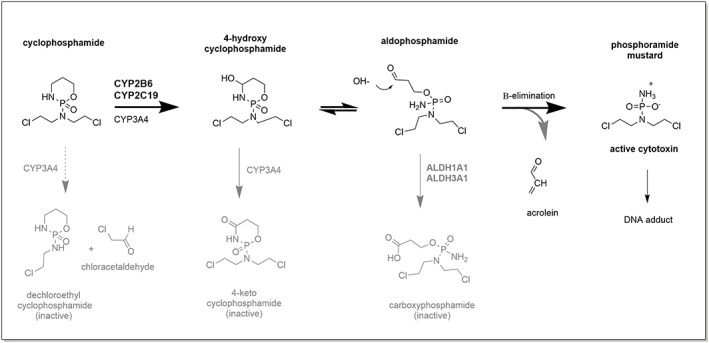
The complex activation and inactivation pathways of cyclophosphamide. These pathways involve enzymes such as the cytochrome P450 (CYP) and aldehyde dehydrogenase (ALDH). Only the major metabolic products are shown, with the inactivation pathways shown in grey. The major route of biotransformation of cyclophosphamide is via 4‐hydroxylation6 whereas CYP3A4 catalysed N‐dechloroethylation is a minor route contributing on average 19% of metabolic clearance7
4‐OHCP exists in equilibrium with its tautomer aldophosphamide in the systemic circulation.17, 18 It is not known how either 4‐OHCP or aldophosphamide enter cells; transport is widely assumed to occur via passive diffusion; however, direct evidence of this is lacking. Once inside cells, aldophosphamide is thought to undergo spontaneous hydrolysis by nonenzymatic β‐elimination, giving rise to the active alkylating compound phosphoramide mustard and the by‐product acrolein. The involvement of phosphodiesterase enzymes in this final activation step has been postulated.19 Phosphoramide mustard is a potent DNA alkylating agent that can readily form DNA interstrand cross links (ICL) via 2 reactive chloroethylamine groups. Phosphoramide mustard reacts with the N7 of guanine on opposite strands of DNA and is highly selective for 5′‐GNC‐3′ sequences.20 These ICL block both DNA replication and gene transcription, resulting in cellular apoptosis. Proliferating cells are most susceptible to this genotoxicity.
A number of inactivation pathways also influence the ultimate levels of phosphoramide mustard (reviewed in1). Cyclophosphamide can undergo N‐dechloroethylation, catalysed predominantly by CYP3A4, resulting in the formation of an inactive metabolite 2‐dechloroethylcyclophosphamide and equimolar amounts of the by‐product chloroacetylaldehyde.7 This minor pathway accounts for around 19% of total metabolic clearance of cyclophosphamide. In addition, 4‐OHCP can undergo secondary metabolism by CYP3A4 to form inactive 4‐keto‐cyclophosphamide. The intermediate product in this reaction (imino‐cyclophosphamide) may also undergo glutathione‐conjugation. Alcohol dehydrogenase and aldo‐keto reductase are also thought to play a minor role in the elimination of aldophosphamide by reduction to alcophosphamide.1 In contrast, oxidation of aldophosphamide into carboxyphosphamide, catalysed by ALDH1A1 and ALDH3A1, is an important inactivation pathway.21, 22, 23 Indeed carboxyphosphamide is a major metabolite of cyclophosphamide.24 Importantly high ALDH activity is expected to protect cells, for example haematopoietic progenitor (CD34+) cells, from the cytotoxic effect of 4‐OHCP.25, 26 This is pertinent for the use of cyclophosphamide as a conditioning agent for haematological stem cell transplantation and in the treatment of autoimmune diseases.
It is clear that there are numerous pathways that could influence interindividual variability of cyclophosphamide response. However, most pharmacogenomic studies have focussed on assessment of the genetic variants of CYP2B6 and CYP2C19, the enzymes that catalyse the initial bioactivation step. The expression and activity of both CYP2B6 and CYP2C19 are highly polymorphic due to common single nucleotide polymorphisms (SNPs).27, 28 The nomenclature of the most commonly studied SNP in these 2 enzymes in the context of cyclophosphamide pharmacogenetics are summarised in Table 1. The coding region SNP in CYP2B6 alter the amount or activity of enzyme produced, which can be substrate dependent (reviewed in28, 29). For example, the 516G>T SNP (rs3745274) in recombinant systems increased enzyme activity due to homotrophic co‐cooperativity30; however, this SNP also causes aberrant splicing, resulting in mRNA lacking exons 4–6 and lower amounts of functional protein.31 CYP2B6 pharmacogenetics are complex because the common protein coding SNP can exist in various haplotype combinations. Notably, the *6 allele (rs2279343 + rs3745274 haplotype) is most prevalent in populations with European ancestry. In contrast, the coding region SNP in CYP2C19 result in no‐function alleles, with the *2 allele most common in people of European ancestry and the *3 allele also prevalent in people of Asian ancestry.
Table 1.
The CYP2C19 and CYP2B6 polymorphisms commonly studied in cyclophosphamide pharmacogenetics
| Gene | SNP | Variant (or definitive haplotype) | Allele | Effecta |
|---|---|---|---|---|
| CYP2C19 | rs4244285 | 681G>A | *2 | Null function |
| rs4986893 | 636G>A | *3 | Null function | |
| CYP2B6 | rs2279343 | 785A>G | *4 | Altered function and expression |
| rs3211371 | 1459C>T | *5 | Altered function and expression | |
| (516G>T and 785A>G) | *6 | Altered function and expression | ||
| (516G>T, 785A>G and 1459C>T) | *7 | Altered function and expression | ||
| rs3745274 | 516G>T | *9 | Altered function and expression | |
| rs4802101 | –750T>C | *G | Alters an HNF transcription factor binding site (HNF1) |
In vitro studies indicate that the functional effects of CYP2B6 variants appear to be substrate dependent.29 SNP, single nucleotide polymorphism.
Regulation of the transcription of CYP2B6 and CYP2C19 is controlled by a number of transcription factors which include: constitutive androstane receptor (CAR), pregnane X receptor (PXR), glucocorticoid receptor (GR), estrogen receptor‐α and GATA‐4.27, 28, 32, 33, 34 CYP2B6 is a highly inducible enzyme28 and can undergo autoinduction by cyclophosphamide.35 Whilst CYP2C19 is also inducible27 the effect of cyclophosphamide on the regulation of this gene is not known.
2. PLASMA PHARMACOKINETICS
There is substantial variation in the plasma pharmacokinetics of cyclophosphamide and total clearance ranges from 1.0–12.6 L/h.6, 36, 37, 38 The major route of clearance is nonrenal and 4‐hydroxylation is the predominant route of hepatic metabolism of the drug. Cyclophosphamide can induce its own metabolism within 24 hours following continuous infusion or after repeated administration over several days.38
In adult cancer patients, CYP2C19 genotype, but not CYP2B6, is significantly associated with cyclophosphamide elimination rate (Ke), but this was only observed in patients receiving ≤1000 mg/m2 dose.39 Population‐pharmacokinetic modelling of data from adult patients (n = 124) receiving 4 day cyclophosphamide regimens, found a trend between CYP2C19*2 status and decreased induced clearance (via 4‐hydroxylation) with an effect size of 14%.40 This regimen included thiotepa, a CYP2B6 inhibitor, which will have influenced the relative contribution of CYP2B6 in cyclophosphamide clearance in these patients. However, inclusion of CYP2C19 and CYP2B6 covariates did not influence the POP‐PK model of clearance (induced) in data from 21 patients undergoing myeloablation with high dose cyclophosphamide. It is not clear which of the CYP2B6 SNPs were assessed in this study.41
Paediatric use of cyclophosphamide for malignancies such as non‐Hodgkin's lymphoma, acute lymphoblastic leukaemia, neuroblastoma and rhabdomyosarcoma suggests that plasma half‐life is shorter in children than adults.37 This is assumed to be due to higher metabolic clearance in children.6 A study in 51 children (median age 5.5 years) did not find a correlation between cyclophosphamide clearance and CYP2B6 genotype.14 This study did not assess CYP2C19. In contrast, in children (mean age 11.2 ± 4 years) with non‐Hodgkins lymphoma (n = 49) an association was observed between the CYP2B6*6 allele and lower clearance of cyclophosphamide both at dose 1 and at dose 5 (after autoinduction).42 No association was found in this study with CYP2C19*2. It is of note that hepatic CYP2B6 enzyme appears to reach adult activity levels as early as 1 year postnatal age.43 In contrast, maturation of hepatic CYP2C19 enzyme activity does not occur until around 10 years of age.44 Hence, assessment of CYP2C19 genotype in very young children is unlikely to uncover any associations with cyclophosphamide clearance.
The plasma pharmacokinetics of 4‐OHCP are also highly variable, and there is a strong correlation between cyclophosphamide and 4‐OHCP plasma AUC.45 The bioactivation ratio, the ratio of 4‐OHCP to the parent drug, is often used as a descriptor of this relationship. Using this bioactivation ratio approach, an association with CYP2C19, but not CYP2B6 genotype was demonstrated in 68 breast cancer patients.46 In contrast, a study in a cohort of 103 cancer patients found no significant association between the bioactivation ratio and either CYP2C19 or variants of CYP2B6.47 Assessment of the bioactivation ratio in a paediatric population (n = 51) also found no association with CYP2B6 genotype; however, CYP2C19 variants were not assessed.14 Most notably, assessment of 4‐OHCP plasma pharmacokinetics at first dose in a large study (n = 567) of non‐Hodgkin's lymphoma patients demonstrated strong (P < 0.0001) independent associations of both CYP2B6 and CYP2C19*2 genotype with 4‐OHCP AUC.48
The relationship between plasma pharmacokinetics and these pharmacogenes has also been studied in patients with (systemic lupus erythematosus, SLE) and vasculitis. A pharmacokinetic study in 23 patients with these autoimmune diseases demonstrated that carriers of CYP2B6 516G>T had a significantly lower cyclophosphamide elimination rate (Ke), however, this study did not assess CYP2C19 genotype.49 These autoimmune diseases can result in glomerulonephritis (i.e. lupus nephritis). A preliminary assessment in a small group of lupus nephritis patients (n = 16) suggested that the combination genotype based on these 2 pharmacogenes may relate to the bioactivation ratio.15 A further study assessed cyclophosphamide and 4‐OHCP pharmacokinetics in 18 lupus nephritis patients did not find an association with CYP2C19 genotype50 but did not determine CYP2B6 genotype. However, a large study (189 SLE patients) found strong correlations between both CYP2C19 and CYP2B6 variants and 4‐OHCP plasma concentrations.51 Multivariate linear regression indicated that CYP2C19*2 genotype accounted for 23.6% of variation in plasma 4‐OHCP. No correlations were observed for the coding region variants of CYP2B6. However, an SNP in the promoter region of the CYP2B6 gene, (−750T>C;*G, rs4802101), was found to be important. A combined genotype determined across the 2 gene loci (CYP2C19 and CYP2B6) strongly associated with plasma 4‐OHCP concentrations and accounted for 47.9% of the individual variability. It is of note that the 5′ promoter region variants, −750T>C (*G) and −2320T>C, were also found to have significant correlations with the bioactivation ratio in a cohort of 103 patients with lymphoma and breast cancer.47 Since CYP2B6 is a highly inducible gene, these promoter region variants may influence the known ability of cyclophosphamide to autoinduce its own metabolism.1, 35, 52
Only a small number of studies have assessed the relationship between pharmacogenetics and the inactivation pathways. Formation of the inactive metabolites (keto‐cyclophosphamide, carboxy‐cyclophosphamide or dechloroethyl‐cyclophosphamide) did not associate with CYP2B6 variants or CYP2C19*2 in 49 children with non‐Hodgkins lymphoma.42 Assessment of these same inactivated metabolites in 51 breast cancer patients also did not find any statistical associations with CYP2B6 or CYP2C19 after Bonferroni correction for multiple testing.53 Formation of 4‐OHCP was not determined in these studies. In contrast, a gene–dose‐dependent association of ALDH3A1 with increased bioactivation ratio has been reported. Notably, this effect is in the opposite direction to the association between CYP2C19 genotype and bioactivation ratio in the same patients.46
Thus, whilst there are substantial inconsistencies in the design and power of each individual study, there does appear to be substantial evidence of an association between both CYP2C19 and CYP2B6 genetic variation and interindividual differences in the bioactivation of cyclophosphamide to 4‐OHCP. These studies are summarised in Table 2. However, the time‐to‐maturation of CYP2C19 (for paediatric studies), use of CYP2B6 inhibitors in concomitant chemotherapy regimens as well as the autoinduction effect of high dose/continuous dosing schedules, which are likely to influence CYP2B6 expression, may be confounding factors in some of these studies. The possible additional influence of ALDH3A1 on the plasma concentrations of 4‐OHCP has not been well characterised.
Table 2.
Summary of studies assessing the relationship between CYP2B6 and CYP2C19 genetic polymorphisms and cyclophosphamide bioactivation plasma pharmacokinetics
| Disease | Study size (n) | Variant allele assessedb | Significant association | |||||
|---|---|---|---|---|---|---|---|---|
| Population ancestrya | CYP2B6 SNP (haplotypes assessed) | CYP2C19 | Multivariate analysis | CYP2B6 | CYP2C19 | |||
| Cancer | 14 | 51 | 516G>T, 785A>G, 1459C>T (*4, *5, *6) | ND | No | ‐ | ND | |
| 39 | 60 | 64C>T, 516G>T, 777C>A, 785A>G, 1459C>T (*2, *3, *4, *5, *6, *7) | *2 | No | ‐ | * 2 | ||
| 42 | 49 | 516G>T, 785A>G, 1459C>T (*5, *6) | *2, *17 | Yes | * 5, * 6 | ‐ | ||
| 40 | 124 | 64C>T, 516G>T, 785A>G, 1459C>T | *2 | Yes | ‐ | ‐ | ||
| 45 | 29 | 516G>T (*9) | *2 | No | * 9 | ‐ | ||
| 46 | 68 | Pakistan | 516G>T, 785A>G, 1459C>T (*4, *5, *6) | *2 | Yes | * 4, * 5, * 6 | ‐ | |
| 47 | 103 | –2320T>C, −750T>C, 136A>G, 296G>A, 419G>A, 415A>G, 516G>T, 785A>G, 1172T>A, 1459C>T, 15582C>T, 18492T>C (*G, *H, *4, *5, *6, *7, *8, *9, *11, *12, *14, *15) | *2, *3 | No | * H | ‐ | ||
| 48 | 567 | Chinese | 64C>T, 516G>T, 785A>G, 1459C>T (*2, *4, *6, *9, *29, *30) | *2, *3 | Yes | * 2, * 4, * 6, * 9, * 29, * 30 | * 2 | |
| Autoimmune disease | 41, c | 21 | 64C>T, 516G>T, 777C>A, 785A>G (*2, *3, *6) | *2 | Yes | ‐ | ‐ | |
| 15 | 16 | Polynesian | 516G>T, 785A>G, 1459C>T (*4, *5, *6, *7, *9) | *2, *3 | Combined genotype | * 5 | * 2, * 3 | |
| 49 | 23 | African American | 516G>T, 785A>G, 1459C>T (*4, *5, *9) | ND | No | * 9 | ND | |
| 50 | 18 | Indian | ND | *2 | No | ND | ‐ | |
| 51 | 189 | Chinese | –2320T>C, −750T>C, 516G>T, 785A>G, 1459C>T, 15582C>T | *2, *3 | Combined genotype | * G, * H, 15582C>T | * 2 | |
European ancestry was major ethnicity, unless indicated.
Alleles are as described in Table 1 with additional alleles (SNP or haplotypes) as follows: CYP2B6*H (−750T>C and –2320T>C) = rs4802101 and rs7254579; *2 (64C>T) = rs8192709; *3 (777C>T) = rs45482602; *8 (415A>G) = rs12721655; *11 (136A>G) = rs35303484; *12 (296G>A) = rs36060847; *14 (419G>A) = rs35773040; *15 (1172T>A) = rs35979566; *29 and *30 are CYP2B6‐CYP2B7 hybrids (crossover in intron 4)54; CYP2B6 15582C>T = rs4803419, 18492T>C = rs2279345; CYP2C19*17 (–806C>T) = rs12248560.
This study did not fully disclose the CYP2B6 and CYP2C19 SNP assessed. ND, not determined; SNP, single nucleotide polymorphism.
3. THERAPEUTIC OUTCOMES
Several articles have also studied associations with the CYP2B6 and CYP2C19 pharmacogenes and therapeutic outcomes. A recent study of chronic lymphocytic leukaemia patients (n = 44), assessed CYP2B6*6 genotype and found no correlation with disease response.55 In contrast, in multiple myeloma patients (n = 26) treated with high‐dose cyclophosphamide for myeloablation, an association was observed between progression free survival and CYP2B6 (785A>G).56 However, CYP2C19 was not assessed in these patients. A larger study (n = 119) of patients with chronic lymphocytic leukaemia found that CYP2B6*6 carriers had inferior treatment response compared with noncarriers of this haplotype (odds ratio [OR] 0.27, P = 0.004).57 Unfortunately, CYP2C19 was again not assessed in these studies. In lymphoma patients (n = 93) treated with high‐dose cyclophosphamide prior to haematological stem cell transplantation, associations between disease relapse, overall survival and CYP2B6 (1459C>T and the *7 haplotype), but not CYP2C19*2, have been reported.58 However, in a study of acute myelogenous and lymphocytic leukaemia patients (n = 359) receiving high‐dose cyclophosphamide for myeloablation prior to haematological stem cell transplantation, patients homozygous for CYP2C19*2 had significantly worse progression free and overall survival compared with individuals who were heterozygote carriers or wild type at this allele.59 This study also assessed CYP2B6 coding region variants and classified patients by assumed phenotypes (poor metabolisers: CYP2B6*6 homozygous individuals; ultrarapid metabolisers: CYP2B6*4 heterozygote carriers; extensive metabolisers: individuals who were any other genotype). Better progression free survival was observed in poor metabolisers compared with ultrarapid metabolisers. Unfortunately, multivariate analysis of genetic variants at the 2 loci was not undertaken in this study. Most recently, significant associations between treatment response and both CYP2C19*2 and CYP2B6 (785A>G) genotype were observed in a large study of lymphoma patients (n = 567). Patients homozygous for CYP2C19*2 had poor response (OR 0.26, 95% confidence interval [CI] 0.05–0.58). Binary logistic models indicated that patients with both CYP2C19*2 and CYP2B6 785G alleles had the worse treatment response.48
A study of CYP2B6 haplotypes in 38 breast cancer patients found a significant gene‐dose relationship and time to relapse,60 while no association was observed with outcomes and CYP2B6 haplotypes in a larger cohort of breast cancer patients (n = 350).61 However, although this study did not assess CYP2C19, disease‐free survival was associated (P = 0.02, unadjusted) with CYP3A4*1B. Importantly, a comprehensive study of both CYP2C19 and CYP2B6 in breast cancer patients (n = 230) found associations with overall survival and these pharmacogenes.62 CYP2B6 516G>T and A785A>G homozygous individuals were more likely to have poor overall survival (P =0.04 and 0.036, respectively). CYP2C19*2 homozygous individuals had an overall survival hazards ratio of 6.2 (95% CI 0.79–487; P = 0.083) compared with wildtype individuals. Unfortunately, multivariate analysis of the epistatic effect of both CYP2C19 and CYP2B6 was not undertaken. Indeed, a separate study using both multivariate and multifactor dimensionality reduction analysis found strong gene–gene interactions for CYP2C19 and CYP2B6 with treatment response in 111 breast cancer patients.63
Of note, a recent global screening (Illumina SNP array) approach confirmed the importance of CYP2C19*2 with disease outcome in 250 Indian breast cancer patients.64 This CYP2C19 SNP (rs 4244285), along with an SNP in ALDH1A1 (rs6151031), was identified as important (p < 5 × 10−8). Also a significant gene–dose effect of CYP2C19*2 with risk of poor response was observed. Of particular note, this genomic approach, which assessed 700 000 SNPs, did not find any association with CYP2B6. However, CYP2B6 may be difficult to accurately assess using this type of genomic analysis due to the presence of the adjacent pseudogene (CYP2B7P1), fusion alleles and copy number variation.54
The relationship between cyclophosphamide pharmacogenetics and outcomes in autoimmune disease has not been as widely studied. Assessment of lupus nephritis patients (n = 62) found that homozygosity for CYP2C19*2 and CYP2B6 (1459C>T) was associated with increased risk of developing end stage renal disease after treatment with cyclophosphamide.65 In contrast, in another study of lupus nephritis patients (n = 36), there was no association between CYP2C19*2 or CYP2B6 (1459C>T) and disease remission.66 However, this study did not assess other CYP2B6 haplotypes that contribute to bioactivation. An additional study of lupus nephritis patients (n = 70) also found no association with CYP2C19 or CYP2B6 and remission from disease. Instead, a polymorphism in GSTP1 (105 Ile > Val) was identified as possibly important (P =0.047).67 Most recently, a combined genetic marker (CYP2C19*2 and CYP2B6‐750C) significantly associated with worse disease remission and longer response time in a gene‐dose dependent manner in SLE patients (n = 189).51 A further study (n = 136) of lupus nephritis patients also found that CYP2C19*2 genotype associated with poor response.50 Additional epistatic effects of CYP3A4 and GSTP1 were suggested. However, logistic regression analysis indicated that CYP2C19*2 genotype independently associated with response (Coeff 0.99 ± SE 0.34, OR 2.69 (95% CI 1.36–5.32), P =0 .0043). Unfortunately, CYP2B6 variants were not assessed in this study.
In a study of patients with renal disease due to vasculitis (n = 93), no association between response and CYP2C19*2 or CYP2B6 (1459C>T) genotype was observed.68 Other important CYP2B6 variants and haplotypes were not assessed. This study did find an association between disease outcome and Fcγ receptor gene (FCGR). Further to this, an Illumina array based analysis of 491 617 SNPs in lupus nephritis patients (n = 109) also found that treatment response at 6 months associated with the FCGR2B locus.69 Hence, renal remission outcomes in these autoimmune diseases may be more strongly influenced by the pathology of the disease (autoimmune reaction to IgG‐complexes) rather than the drug treatment. However, this genome array‐based approach, as noted earlier in this review, may not report on CYP2B6 SNP due to the complexity of this genomic region. The studies reporting associations of CYP2B6 and/or CYP2C19 genetic polymorphisms with therapeutic outcomes are summarised in Table 3.
Table 3.
Summary of studies assessing the relationship between CYP2B6 and CYP2C19 genetic polymorphisms and therapeutic outcomes
| Disease | Study size (n) | Population ancestrya | Variant allele assessedb | Significant association | ||||
|---|---|---|---|---|---|---|---|---|
| CYP2B6 SNP (haplotypes assessed) | CYP2C19 | Multivariate analysis | CYP2B6 | CYP2C19 | ||||
| Cancer | 48 | 567 | Chinese | 64C>T, 516G>T, 785A>G, 1459C>T (*2, *4, *6, *9, *29, *30) | *2, *3 | Yes | * 4 | * 2 |
| 55 | 44 | 516G>T, 785A>G (*6) | ND | No | ‐ | ND | ||
| 56 | 26 | 516G>T, 785A>G, 1459C>T (*4, *5, *6, *9) | ND | No | * 4 | ND | ||
| 57 | 119 | 516G>T, 785A>G (*6) | ND | No | * 6 | ND | ||
| 58 | 93 | 516G>T, 785A>G, 1459C>T (*5, *6, *7) | *2 | Yes | * 5, * 7 | ‐ | ||
| 59, c | 359 | 516G>T, 785A>G, 1459C>T (*4, *5, *6, *7) | *2, *3 | No | * 6 | * 2, * 3 | ||
| 60, c | 38 | Middle Eastern | 516G>T, 785A>G, 1459C>T (*4, *5, *6, *9) | ND | No | * 5, * 6, * 9 | ND | |
| 61 | 350 | 516G>T, 785A>G, 1459C>T (*4, *5, *6, *7, *9) | ND | Yes | ‐ | ND | ||
| 62 | 230 | 64C>T, 415A>G, 516G>T, 777C>A, 785A>G, 1459C>T (*2, *3, *4, *5, *8, *9) | *2 | No | * 2, * 4, * 8, * 9 | * 2 | ||
| 63 | 111 | Indian | 516G>T (*9) | *2 | Yes | |||
| 64 | 250 | Indian | GWAS (700 000 SNP) | ‐ | ‐ | * 2 | ||
| Autoimmune disease | 50 | 136 | Indian | ND | *2 | Yes | ND | * 2 |
| 51, d | 189 | Chinese | –750T>C, −2320T>C, 785A>G, 1459C>T, 516G>T, 15582C>T | *2, *3 | Combined genotype | CYP2B6 * H – and –CYP2C19 * 2 | ||
European ancestry was major ethnicity, unless indicated.
Alleles are as described in Table 1 with the additional alleles (SNP or haplotypes) as follows: CYP2B6*H (−750T>C and –2320T>C) = rs4802101 and rs7254579; *2 (64C>T) = rs8192709; *3 (777C>T) = rs45482602; *8 (415A>G) = rs12721655; *11 (136A>G) = rs35303484; *12 (296G>A) = rs36060847; *14 (419G>A) = rs35773040; *15 (1172T>A) = rs35979566; *29 and *30 are CYP2B6‐CYP2B7 hybrids (crossover in intron 4)54; 15582C>T = rs4803419; CYP2C19*17 (–806C>T) = rs12248560.
These studies stratified patients by a putative metaboliser status based on various combinations of CYP2B6 alleles assessed.
This study stratified patients by a putative metaboliser status based on combinations of both CYP2B6 and CYP2C19 alleles assessed. GWAS, genome‐wide association study; ND, not determined; SNP, single nucleotide polymorphism.
Despite the inconsistent assessment of both CYP2B6 and CYP2C19 in many of the studies reviewed supra vide there does appear to be substantial evidence for the role of both CYP2B6 and CYP2C19 pharmacogenes with cyclophosphamide response. It is important to note that these associations were observed in many different contexts despite the use of different dosages, dosing regimens, drug combinations, as well as differences in the intrinsic resistance in different neoplastic and autoimmune diseases. However, the different statistical methods and gene analysis methods employed across these studies may hide or enhance associations.
It is clear that the majority of the therapeutic outcome or pharmacokinetic studies have focussed on CYP2B6 and/or CYP2C19 as candidate genes. However, some studies have also reported on selected SNP in other candidate genes such as CYP2C9, GST or ABCB1.39, 40, 41, 42, 46, 48, 49, 50, 51, 55, 56, 61, 62, 63, 64, 65, 67, 68 Whilst not the focus of this review, it is important to note that the role of pharmacogenetics in the risk of excessive toxicity (due to high levels of bioactivation of this cytotoxic drug) have also been assessed. Associations between CYP2C19 and/or CYP2B6, as well as other pharmacogenes such as ALDH, GSTP1 and CYP3A4, and the severity of neutropenia or premature ovarian failure have been reported.40, 65, 67, 70, 71, 72, 73, 74, 75
4. CONCLUSION
It appears from pharmacokinetic studies that both CYP2C19 and CYP2B6 influence the bioactivation of cyclophosphamide to 4‐OHCP. Inherited variation in both of these genes also appear to influence therapeutic outcomes. However, many studies do not comprehensively investigate both of these pharmacogenes and this has led to inconsistencies in the quality of the data. Approximately 25% of the reported studies have not assessed CYP2C19 loss of function variants and in the light of the recent large studies,48, 51, 62 which have demonstrated significant associations with the CYP2C19*2 null function variant, future studies should include assessment of this locus. Consideration of individuals who are compound heterozygous for null function alleles (*2/*3) should also be incorporated in these studies, particularly in non‐European populations. This appears to have been overlooked48, 51 despite the high prevalence of the *3 allele in these populations.
Comprehensive assessment of the multiple SNP variants in CY2B6 is also variable across the 28 studies reviewed. CYP2B6 is a more challenging genomic region than CYP2C19. CYP2B6 is a highly polymorphic locus and the correct assignment of allele (*star number) is important, since there appear to be substantial substrate‐dependent differences in the in vitro activity of each variant and haplotype combination (reviewed in29). The 516G>T variant is in strong linkage disequilibrium (>0.8) with the 785A>G variant, hence the *6 haplotype is common. However, it is important to note that both of these variants can also be found in isolation (*4 or *9 allele). Most studies in this review have not directly assessed the chromosome location (maternal or paternal) of the SNP variant (i.e. diplotype). Thus, for example, individuals who are carriers of both 516G>T and 785A>G variants could be either heterozygous for the *6 allele (diplotype: GA/TG), or compound heterozygous (*4/*9) with a diplotype of GG/TA. Statistical methods for imputation of the most likely haplotype (e.g. PHASE algorithm76) could be incorporated in future studies; however, this approach may still incorrectly assign a proportion of the diplotype calls.
CYP2B6 is located adjacent to a highly homologous pseudogene (CYP2B7P1) and duplication and deletion fusion alleles between CYP2B6‐2B7P1 (*29 and *30) also influence cyclophosphamide bioactivation.48 The complexity of this genomic region and the presence of the CYP2B6 SNP (e.g. 785A>G) in the pseudogene can result in poor data quality when using next generation sequencing or genome‐wide SNP arrays. This may affect the routine reporting of this region in this genomic methods and could explain why despite the substantial signal from candidate gene studies (Table 2 and 3) the 2 genome‐wide association studies reviewed64, 69 did not detect the CYP2B6 locus as important for treatment outcomes. Long read amplification of this region is required to accurately call haplotype in heterozygous carriers of common coding region SNP of CYP2B6. Newer techniques, such as high‐resolution melt analysis, multiplex‐ligation‐probe amplification and nanopore sequencing,77, 78, 79 will probably be required to appropriately diplotype patients for CYP2B6 variants and fully assess the role of this pharmacogene in cyclophosphamide outcomes.
Finally, autoinduction of cyclophosphamide bioactivation, following high dose or continuous dosing regimens, is likely to have an important influence on the pharmacogenetic relationships with cyclophosphamide bioactivation. Notably, some studies46, 50 have found associations with SNPs in the promoter region of CYP2B6. These SNPs are thought to alter the binding of HNF1 and HNF4 transcription factors to the CYP2B6 promoter80 and could influence an individual's susceptibility to autoinduction. Assessment of the relationships between CYP2B6 promoter variants and cyclophosphamide bioactivation pharmacokinetics at the initial, as well as subsequent doses, could clarify the importance of these regulatory SNPs.
Future studies to confirm the role of the CYP2B6 and CYP2C19 pharmacogenes should include investigation of epistatic gene–gene interactions using appropriate statistical analysis such as multifactor dimensionality reduction or reporting individual patients combined genotype at the 2 gene loci. This multivariate approach has already shown some utility in population sizes between 189 and 567 patients.50, 51 Remarkably, considering the important role in the inactivation of 4‐OHCP, few studies have included the assessment of ALDH1A1 on clinical response to cyclophosphamide therapy. This detoxification pathway is likely to influence the amount of circulating 4‐OHCP‐aldophosphamide available for uptake into cells and thereby influence the number of DNA ICL formed within cells, and thus patient outcomes. This candidate gene should be included in future studies of cyclophosphamide pharmacogenetics. Using either a comprehensive candidate gene approach or by suitable genome‐wide assessment, understanding the inherited factors that influence response to this important drug may allow appropriate treatment stratification based on an individual patient's combined genotype at these loci.
COMPETING INTERESTS
The authors have no conflicts of interest to declare.
ACKNOWLEDGEMENTS
All authors contributed to the collation of information, drafting and revision of the manuscript. All authors approved the final version of the manuscript.
Helsby NA, Yong M, van Kan M, de Zoysa JR, Burns KE. The importance of both CYP2C19 and CYP2B6 germline variations in cyclophosphamide pharmacokinetics and clinical outcomes. Br J Clin Pharmacol. 2019;85:1925–1934. 10.1111/bcp.14031
REFERENCES
- 1. Zhang J, Tian Q, Chan SY, et al. Metabolism and transport of oxazaphosphorines and the clinical implications. Drug Metab Rev. 2005;37(4):611‐703. [DOI] [PubMed] [Google Scholar]
- 2. Brummaier T, Pohanka E, Studnicka‐Benke A, Pieringer H. Using cyclophosphamide in inflammatory rheumatic diseases. Eur J Intern Med. 2013;24(7):590‐596. [DOI] [PubMed] [Google Scholar]
- 3. Emadi A, Jones RJ, Brodsky RA. Cyclophosphamide and cancer: golden anniversary. Nat Rev Clin Oncol. 2009;6(11):638‐647. [DOI] [PubMed] [Google Scholar]
- 4. Al‐Homsi AS, Roy TS, Cole K, Feng Y, Duffner U. Post‐transplant high‐dose cyclophosphamide for the prevention of graft‐versus‐host disease. Biol Blood Marrow Transplant. 2015;21(4):604‐611. [DOI] [PubMed] [Google Scholar]
- 5. Mathrani V, Alejmi A, Griffin S, Roberts G. Intravenous cyclophosphamide and oral prednisolone is a safe and effective treatment option for idiopathic membranous nephropathy. Clin Kidney J. 2017;10(4):450‐454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. de Jonge ME, Huitema ADR, Rodenhuis S, Beijnen JH. Clinical pharmacokinetics of cyclophosphamide. Clin Pharmacokinet. 2005;44(11):1135‐1164. [DOI] [PubMed] [Google Scholar]
- 7. Huang Z, Roy P, Waxman DJ. Role of human liver microsomal CYP3A4 and CYP2B6 in catalyzing N‐dechloroethylation of cyclophosphamide and ifosfamide. Biochem Pharmacol. 2000;59(8):961‐972. [DOI] [PubMed] [Google Scholar]
- 8. Chang TKH, Weber GF, Crespi CL, Waxman DJ. Differential activation of cyclophosphamide and ifosphamide by cytochromes P‐450 2B and 3A in human liver microsomes. Cancer Res. 1993;53(23):5629‐5637. [PubMed] [Google Scholar]
- 9. Xie H‐J, Yasar Ü, Lundgren S, et al. Role of polymorphic human CYP2B6 in cyclophosphamide bioactivation. Pharmacogenomics J. 2003;3(1):53‐61. [DOI] [PubMed] [Google Scholar]
- 10. Roy P, Yu LJ, Crespi CL, Waxman DJ. Development of a substrate‐activity based approach to identify the major human liver P‐450 catalysts of cyclophosphamide and ifosfamide activation based on cDNA‐expressed activities and liver microsomal P‐450 profiles. Drug Metab Dispos. 1999;27(6):655‐666. [PubMed] [Google Scholar]
- 11. Chang TK, Yu L, Goldstein JA, Waxman DJ. Identification of the polymorphically expressed CYP2C19 and the wild‐type CYP2C9‐ILE359 allele as low‐km catalysts of cyclophosphamide and ifosfamide activation. Pharmacogenetics. 1997;7(3):211‐221. [DOI] [PubMed] [Google Scholar]
- 12. Griskevicius L, Yasar Ü, Sandberg M, et al. Bioactivation of cyclophosphamide: the role of polymorphic CYP2C enzymes. Eur J Clin Pharmacol. 2003;59(2):103‐109. [DOI] [PubMed] [Google Scholar]
- 13. El‐Serafi I, Fares M, Abedi‐Valugerdi M, et al. Cytochrome P450 2J2, a new key enzyme in cyclophosphamide bioactivation and a potential biomarker for hematological malignancies. Pharmacogenomics J. 2015;15(5):405‐413. [DOI] [PubMed] [Google Scholar]
- 14. Raccor BS, Claessens AJ, Dinh JC, et al. Potential contribution of cytochrome P450 2B6 to hepatic 4‐hydroxycyclophosphamide formation in vitro and in vivo. Drug Metab Dispos. 2012;40(1):54‐63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Helsby NA, Hui C‐Y, Goldthorpe MA, et al. The combined impact of CYP2C19 and CYP2B6 pharmacogenetics on cyclophosphamide bioactivation. Br J Clin Pharmacol. 2010;70(6):844‐853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kishino Y, Hasegawa T, Kato A, et al. Effect of inter‐individual variability in human liver cytochrome P450 isozymes on cyclophosphamide‐induced micronucleus formation. Mutat Res Toxicol Environ Mutagen. 2019;838:37‐45. [DOI] [PubMed] [Google Scholar]
- 17. Fenselau C, Kan M‐NN, Rao SS, Myles A, Friedman OM, Colvin M. Identification of aldophosphamide as a metabolite of cyclophosphamide in vitro and in vivo in humans. Cancer Res. 1977;37(8 Part 1):2538‐2543. [PubMed] [Google Scholar]
- 18. Sladek NE. Metabolism of oxazaphosphorines. Pharmacol Ther. 1988;37(3):301‐355. [DOI] [PubMed] [Google Scholar]
- 19. Voelcker G. Enzyme catalyzed decomposition of 4‐hydroxycyclophosphamide. Open Conf Proc J. 2017;8(1):44‐51. 10.2174/2210289201708010044 [DOI] [Google Scholar]
- 20. Struck RF, Davis RL, Berardini MD, Loechler EL. DNA guanine‐guanine crosslinking sequence specificity of isophosphoramide mustard, the alkylating metabolite of the clinical antitumor agent ifosfamide. Cancer Chemother Pharmacol. 2000;45(1):59‐62. [DOI] [PubMed] [Google Scholar]
- 21. Moreb JS, Muhoczy D, Ostmark B, Zucali JR. RNAi‐mediated knockdown of aldehyde dehydrogenase class‐1A1 and class‐3A1 is specific and reveals that each contributes equally to the resistance against 4‐hydroperoxycyclophosphamide. Cancer Chemother Pharmacol. 2007;59(1):127‐136. [DOI] [PubMed] [Google Scholar]
- 22. Sládek NE, Kollander R, Sreerama L, Kiang DT. Cellular levels of aldehyde dehydrogenases (ALDH1A1 and ALDH3A1) as predictors of therapeutic responses to cyclophosphamide‐based chemotherapy of breast cancer: a retrospective study. Cancer Chemother Pharmacol. 2002;49(4):309‐321. [DOI] [PubMed] [Google Scholar]
- 23. von Eitzen U, Meier‐Tackmann D, Agarwal DP, Goedde HW. Detoxification of cyclophosphamide by human aldehyde dehydrogenase isozymes. Cancer Lett. 1994;76(1):45‐49. [DOI] [PubMed] [Google Scholar]
- 24. Joqueviel C, Martino R, Gilard V, Malet‐Martino M, Canal P, Niemeyer U. Urinary excretion of cyclophosphamide in humans, determined by Phosphorus‐31 nuclear magnetic resonance spectroscopy. Drug Metab Dispos. 1998;26(5):418‐428. [PubMed] [Google Scholar]
- 25. Russo, Hilton J, Colvin OM. The role of aldehyde dehydrogenase isozymes in cellular resistance to the alkylating agent cyclophosphamide. Prog Clin Biol Res. 1989;290:65‐79. [PubMed] [Google Scholar]
- 26. Kastan MB, Schlaffer E, Russo JE, Colvin OM, Civin CI, Hilton J. Direct demonstration of elevated aldehyde dehydrogenase in human hematopoietic progenitor cells. Blood. 1990;75(10):1947‐1950. [PubMed] [Google Scholar]
- 27. Helsby NA, Burns KE. Molecular mechanisms of genetic variation and transcriptional regulation of CYP2C19. Front Genet. 2012;3:206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zanger UM, Klein K. Pharmacogenetics of cytochrome P450 2B6 (CYP2B6): advances on polymorphisms, mechanisms, and clinical relevance. Front Genet[Internet]. 2013;4:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Helsby NA, Tingle MD. Which CYP2B6 variants have functional consequences for cyclophosphamide bioactivation? Drug Metab Dispos. 2012;40(3):635‐637. [DOI] [PubMed] [Google Scholar]
- 30. Ariyoshi N, Miyazaki M, Toide K, Sawamura Y, Kamataki T. A single nucleotide polymorphism of CYP2B6 found in Japanese enhances catalytic activity by autoactivation. Biochem Biophys Res Commun. 2001;281(5):1256‐1260. [DOI] [PubMed] [Google Scholar]
- 31. Hofmann MH, Blievernicht JK, Klein K, et al. Aberrant splicing caused by single nucleotide polymorphism c.516G>T [Q172H], a marker of CYP2B6*6, is responsible for decreased expression and activity of CYP2B6 in liver. J Pharmacol Exp Ther. 2008;325(1):284‐292. [DOI] [PubMed] [Google Scholar]
- 32. Wang H, Faucette SR, Gilbert D, et al. Glucocorticoid receptor enhancement of pregnane X receptor‐mediated CYP2B6 regulation in primary human hepatocytes. Drug Metab Dispos. 2003;31(5):620‐630. [DOI] [PubMed] [Google Scholar]
- 33. Lo R, Burgoon L, MacPherson L, Ahmed S, Matthews J. Estrogen receptor‐dependent regulation of CYP2B6 in human breast cancer cells. Biochim Biophys Acta. 2010;1799(5‐6):469‐479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lee W, Tirona RG, Kim RB. Localization of the distal enhancer elements conferring synergistic activation of the human CYP2B6 gene by the constitutive androstane receptor and hepatocyte nuclear factor 4α. Clin Pharmacol Ther. 2003;73(2):P60‐P60. [Google Scholar]
- 35. Lindley C, Hamilton G, McCune JS, et al. The effect of cyclophosphamide with and without dexamethasone on cytochrome P450 3A4 and 2B6 in human hepatocytes. Drug Metab Dispos. 2002;30(7):814‐822. [DOI] [PubMed] [Google Scholar]
- 36. Batey MA, Wright JG, Azzabi A, et al. Population pharmacokinetics of adjuvant cyclophosphamide, methotrexate and 5‐fluorouracil (CMF). Eur J Cancer. 2002;38(8):1081‐1089. [DOI] [PubMed] [Google Scholar]
- 37. Yule SM, Boddy AV, Cole M, et al. Cyclophosphamide pharmacokinetics in children. Br J Clin Pharmacol. 1996;41(1):13‐19. [DOI] [PubMed] [Google Scholar]
- 38. Slattery JT, Kalhorn TF, McDonald GB, et al. Conditioning regimen‐dependent disposition of cyclophosphamide and hydroxycyclophosphamide in human marrow transplantation patients. J Clin Oncol. 1996;14(5):1484‐1494. [DOI] [PubMed] [Google Scholar]
- 39. Timm R, Kaiser R, Lötsch J, et al. Association of cyclophosphamide pharmacokinetics to polymorphic cytochrome P450 2C19. Pharmacogenomics J. 2005;5(6):365‐373. [DOI] [PubMed] [Google Scholar]
- 40. Ekhart C, Doodeman VD, Rodenhuis S, Smits PHM, Beijnen JH, Huitema ADR. Influence of polymorphisms of drug metabolizing enzymes (CYP2B6, CYP2C9, CYP2C19, CYP3A4, CYP3A5, GSTA1, GSTP1, ALDH1A1 and ALDH3A1) on the pharmacokinetics of cyclophosphamide and 4‐hydroxycyclophosphamide. Pharmacogenet Genomics. 2008;18(6):515‐523. [DOI] [PubMed] [Google Scholar]
- 41. Kim I‐W, Yun H, Choi B, et al. Population pharmacokinetics analysis of cyclophosphamide with genetic effects in patients undergoing hematopoietic stem cell transplantation. Eur J Clin Pharmacol. 2013;69(8):1543‐1551. [DOI] [PubMed] [Google Scholar]
- 42. Veal GJ, Cole M, Chinnaswamy G, et al. Cyclophosphamide pharmacokinetics and pharmacogenetics in children with B‐cell non‐Hodgkin's lymphoma. Eur J Cancer. 2016;55:56‐64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pearce RE, Gaedigk R, Twist GP, et al. Developmental expression of CYP2B6: a comprehensive analysis of mRNA expression, protein content and bupropion hydroxylase activity and the impact of genetic variation. Drug Metab Dispos. 2016;44(7):948‐958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Koukouritaki SB, Manro JR, Marsh SA, et al. Developmental expression of human hepatic CYP2C9 and CYP2C19. J Pharmacol Exp Ther. 2004;308(3):965‐974. [DOI] [PubMed] [Google Scholar]
- 45. Xie H, Griskevicius L, Ståhle L, et al. Pharmacogenetics of cyclophosphamide in patients with hematological malignancies. Eur J Pharm Sci. 2006;27(1):54‐61. [DOI] [PubMed] [Google Scholar]
- 46. Afsar NA, Ufer M, Haenisch S, et al. Relationship of drug metabolizing enzyme genotype to plasma levels as well as myelotoxicity of cyclophosphamide in breast cancer patients. Eur J Clin Pharmacol. 2012;68(4):389‐395. [DOI] [PubMed] [Google Scholar]
- 47. Nakajima M, Komagata S, Fujiki Y, et al. Genetic polymorphisms of CYP2B6 affect the pharmacokinetics/pharmacodynamics of cyclophosphamide in Japanese cancer patients. Pharmacogenet Genomics. 2007;17(6):431‐445. [DOI] [PubMed] [Google Scholar]
- 48. Shu W, Chen L, Hu X, et al. Cytochrome P450 genetic variations can predict mRNA expression, cyclophosphamide 4‐hydroxylation, and treatment outcomes in Chinese patients with non‐Hodgkin's lymphoma. J Clin Pharmacol. 2017;57(7):886‐898. [DOI] [PubMed] [Google Scholar]
- 49. Joy MS, La M, Wang J, et al. Cyclophosphamide and 4‐hydroxycyclophosphamide pharmacokinetics in patients with glomerulonephritis secondary to lupus and small vessel vasculitis. Br J Clin Pharmacol. 2012;74(3):445‐455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kumaraswami K, Katkam SK, Aggarwal A, et al. Epistatic interactions among CYP2C19*2, CYP3A4 and GSTP1 on the cyclophosphamide therapy in lupus nephritis patients. Pharmacogenomics. 2017;18(15):1401‐1411. [DOI] [PubMed] [Google Scholar]
- 51. Shu W, Guan S, Yang X, et al. Genetic markers in CYP2C19 and CYP2B6 for prediction of cyclophosphamide's 4‐hydroxylation, efficacy and side effects in Chinese patients with systemic lupus erythematosus. Br J Clin Pharmacol. 2016;81(2):327‐340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hassan M, Svensson USH, Ljungman P, et al. A mechanism‐based pharmacokinetic‐enzyme model for cyclophosphamide autoinduction in breast cancer patients. Br J Clin Pharmacol. 1999;48(5):669‐677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Jamieson D, Lee J, Cresti N, et al. Pharmacogenetics of adjuvant breast cancer treatment with cyclophosphamide, epirubicin and 5‐fluorouracil. Cancer Chemother Pharmacol. 2014;74(4):667‐674. [DOI] [PubMed] [Google Scholar]
- 54. Martis S, Mei H, Vijzelaar R, Edelmann L, Desnick RJ, Scott SA. Multi‐ethnic cytochrome‐P450 copy number profiling: novel Pharmacogenetic alleles and mechanism of copy number variation formation. Pharmacogenomics J. 2013;13(6):558‐566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Vukovic V, Karan‐Djurasevic T, Antic D, et al. Association of SLC28A3 gene expression and CYP2B6*6 allele with the response to Fludarabine plus cyclophosphamide in chronic lymphocytic leukemia patients. Pathol Oncol Res, 2019. 10.1007/s12253-019-00613-4 [DOI] [PubMed] [Google Scholar]
- 56. Jakobsen Falk I, Khan MS, Thunell L, Nahi H, Green H. Association of CYP2B6 genotype with survival and progression free survival in cyclophosphamide treated multiple myeloma. J Cancer Ther. 2012;3(1):20‐27. [Google Scholar]
- 57. Johnson GG, Lin K, Cox TF, et al. CYP2B6*6 is an independent determinant of inferior response to fludarabine plus cyclophosphamide in chronic lymphocytic leukemia. Blood. 2013;122(26):4253‐4258. [DOI] [PubMed] [Google Scholar]
- 58. Bachanova V, Shanley R, Malik F, et al. Cytochrome P450 2B6*5 increases relapse after cyclophosphamide‐containing conditioning and autologous transplantation for lymphoma. Biol Blood Marrow Transplant. 2015;21(5):944‐948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Melanson SEF, Stevenson K, Kim H, et al. Allelic variations in CYP2B6 and CYP2C19 and survival of patients receiving cyclophosphamide prior to myeloablative hematopoietic stem cell transplantation. Am J Hematol. 2010;85(12):967‐971. [DOI] [PubMed] [Google Scholar]
- 60. Haroun F, Al‐Shaar L, Habib RH, et al. Effects of CYP2B6 genetic polymorphisms in patients receiving cyclophosphamide combination chemotherapy for breast cancer. Cancer Chemother Pharmacol. 2015;75(1):207‐214. [DOI] [PubMed] [Google Scholar]
- 61. Gor PP, Su HI, Gray RJ, et al. Cyclophosphamide‐ metabolizing enzyme polymorphisms and survival outcomes after adjuvant chemotherapy for node‐positive breast cancer: a retrospective cohort study. Breast Cancer Res BCR. 2010;12(3):R26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Bray J, Sludden J, Griffin MJ, et al. Influence of pharmacogenetics on response and toxicity in breast cancer patients treated with doxorubicin and cyclophosphamide. Br J Cancer. 2010;102(6):1003‐1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Tulsyan S, Agarwal G, Lal P, Mittal B. Significant role of CYP450 genetic variants in cyclophosphamide based breast cancer treatment outcomes: a multi‐analytical strategy. Clin Chim Acta. 2014;434:21‐28. [DOI] [PubMed] [Google Scholar]
- 64. Kalra S, Kaur RP, Ludhiadch A, et al. Association of CYP2C19*2 and ALDH1A1*1/*2 variants with disease outcome in breast cancer patients: results of a global screening array. Eur J Clin Pharmacol. 2018;74(10):1291‐1298. [DOI] [PubMed] [Google Scholar]
- 65. Takada K, Arefayene M, Desta Z, et al. Cytochrome P450 pharmacogenetics as a predictor of toxicity and clinical response to pulse cyclophosphamide in lupus nephritis. Arthritis Rheum. 2004;50(7):2202‐2210. [DOI] [PubMed] [Google Scholar]
- 66. Winoto J, Song H, Hines C, Nagaraja H, Rovin BH. Cytochrome P450 polymorphisms and the response of lupus nephritis to cyclophosphamide therapy. Clin Nephrol. 2011;75(05):451‐457. [DOI] [PubMed] [Google Scholar]
- 67. Audemard‐Verger A, Martin Silva N, Verstuyft C, et al. Glutathione S transferases polymorphisms are independent prognostic factors in lupus nephritis treated with cyclophosphamide. PLoS ONE. 2016;11(3):e0151696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Cartin‐Ceba R, Indrakanti D, Specks U, et al. The pharmacogenomic association of Fcγ receptors and cytochrome P450 enzymes with response to rituximab or cyclophosphamide treatment in antineutrophil cytoplasmic antibody‐associated Vasculitis. Arthritis Rheumatol Hoboken NJ. 2017;69(1):169‐175. [DOI] [PubMed] [Google Scholar]
- 69. Kim K, Bang S‐Y, Joo YB, et al. Response to intravenous cyclophosphamide treatment for lupus nephritis associated with polymorphisms in the FCGR2B‐FCRLA locus. J Rheumatol. 2016;43(6):1045‐1049. [DOI] [PubMed] [Google Scholar]
- 70. Tsuji D, Ikeda M, Yamamoto K, et al. Drug‐related genetic polymorphisms affecting severe chemotherapy‐induced neutropenia in breast cancer patients: a hospital‐based observational study. Medicine (Baltimore). 2016;95(44):e5151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Su HI, Sammel MD, Velders L, et al. Association of cyclophosphamide drug–metabolizing enzyme polymorphisms and chemotherapy‐related ovarian failure in breast cancer survivors. Fertil Steril. 2010;94(2):645‐654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Helsby N, Goldthorpe M, Gow P, de Zoysa J. Oral cyclophosphamide confounds the relationship between CYP2C19 and CYP2B6 pharmacogenetics and cyclophosphamide‐induced premature ovarian failure in lupus nephritis patients. Adv Pharmacol Pharm. 2013;1(1):1‐8. [Google Scholar]
- 73. Schirmer JH, Bremer JP, Moosig F, et al. Cyclophosphamide treatment‐induced leukopenia rates in ANCA‐associated vasculitis are influenced by variant CYP450 2C9 genotypes. Pharmacogenomics. 2016;17(4):367‐374. [DOI] [PubMed] [Google Scholar]
- 74. Black JL, Litzow MR, Hogan WJ, et al. Correlation of CYP2B6, CYP2C19, ABCC4 and SOD2 genotype with outcomes in allogeneic blood and marrow transplant patients. Leuk Res. 2012;36(1):59‐66. [DOI] [PubMed] [Google Scholar]
- 75. Ngamjanyaporn P, Thakkinstian A, Verasertniyom O, et al. Pharmacogenetics of cyclophosphamide and CYP2C19 polymorphism in Thai systemic lupus erythematosus. Rheumatol Int. 2011;31(9):1215‐1218. [DOI] [PubMed] [Google Scholar]
- 76. Stephens M, Donnelly P. A comparison of Bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet. 2003;73(5):1162‐1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Jain M, Koren S, Miga KH, et al. Nanopore sequencing and assembly of a human genome with ultra‐long reads. Nat Biotechnol. 2018;36(4):338‐345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Futatsugawa Y, Kubota T, Ishiguro A, Suzuki H, Ishikawa H, Iga T. PCR‐based haplotype determination to distinguish CYP2B6*1/*7 and *5/*6. Clin Chem. 2004;50(8):1472‐1473. [DOI] [PubMed] [Google Scholar]
- 79. Twist GP, Gaedigk R, Leeder JS, Gaedigk A. High‐resolution melt analysis to detect sequence variations in highly homologous gene regions: application to CYP2B6. Pharmacogenomics. 2013;14(8):913‐922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Lamba V, Lamba J, Yasuda K, et al. Hepatic CYP2B6 expression: gender and ethnic differences and relationship to CYP2B6 genotype and CAR (constitutive Androstane receptor) expression. J Pharmacol Exp Ther. 2003;307(3):906‐922. [DOI] [PubMed] [Google Scholar]


