Abstract
Background
Gliomas are the most frequent and aggressive malignancies of the central nervous system. Decades of molecular analyses have demonstrated that gliomas accumulate genetic alterations that culminate in enhanced activity of receptor tyrosine kinases and downstream mediators. While the genetic alterations, like gene amplification or loss, have been well characterized, little information exists about changes in the proteome of gliomas of different grades.
Methods
We performed unbiased quantitative proteomics of human glioma biopsies by mass spectrometry followed by bioinformatic analysis.
Findings
Various pathways were found to be up- or downregulated. In particular, endocytosis as pathway was affected by a vast and concomitant reduction of multiple machinery components involved in initiation, formation, and scission of endocytic carriers. Both clathrin-dependent and -independent endocytosis were changed, since not only clathrin, AP-2 adaptins, and endophilins were downregulated, but also dynamin that is shared by both pathways. The reduction of endocytic machinery components caused increased receptor cell surface levels, a prominent phenotype of defective endocytosis. Analysis of additional biopsies revealed that depletion of endocytic machinery components was a common trait of various glioma grades and subclasses.
Interpretation
We propose that impaired endocytosis creates a selective advantage in glioma tumor progression due to prolonged receptor tyrosine kinase signaling from the cell surface.
Fund
This work was supported by Grants 316030-164105 (to P. Jenö), 31003A-162643 (to M. Spiess) and PP00P3-176974 (to G. Hutter) from the Swiss National Science Foundation. Further funding was received by the Department of Surgery from the University Hospital Basel.
Keywords: Glioma, Endocytosis, RTK, Dynamin, Clathrin, AP-2
Research in context.
Evidence before this study
Gliomas are the most frequent and the most aggressive malignancies of the central nervous system. Decades of molecular genetic analyses have shown that gliomas accumulate genetic alterations that result in the enhanced activity of growth factor receptor tyrosine kinases (RTKs), and mediators of downstream signaling pathways. Among them are gain-of-function of EGFR, PDGFR, PI3KA or BRAF, and loss-of-function of PTEN or NF1, resulting in exacerbated proliferative responses. While the genetic alterations are well established, little information exists about proteomic changes of gliomas of different grades and subclasses.
Added value of this study
Using unbiased proteomics profiling, we found various pathways to be up- or downregulated in human glioma biopsies of different grades. In particular, endocytosis as pathway was affected by a vast and concomitant depletion of multiple machinery components involved in the generation of clathrin-dependent and -independent endocytic carriers for RTK internalization. In addition, by expanding our analysis to other glioma subclasses, we found that downregulation of endocytosis is a common trait of gliomas. Thus, the reduction of endocytic machinery components might maintain and potentially enhance growth factor signaling from the cell surface.
Implications of all the available evidence
Quantitative proteomics revealed a shutdown of endocytosis machinery components in human glioma to a various extent. Our results suggest that impaired endocytosis is an additional strategy used by gliomas to stimulate cell proliferation. Moreover, the present study also provides an inventory of proteins that are quantitatively regulated in glioma and could be potentially clinically exploited.
Alt-text: Unlabelled Box
1. Introduction
Gliomas are the most frequent and aggressive malignancies of the central nervous system with a median survival rate of 14 months for the World Health Organization (WHO) grade IV glioblastoma (GB) patients [1]. Current adjuvant chemotherapy based on the alkylating agent temozolomide results in a relative survival benefit for glioma patients with methylation of the MGMT gene promoter [2]. A posteriori, gliomas with this molecular feature turned out to include tumors with the glioma CpG island methylator phenotype (G-CIMP), that is tightly associated with mutations at the genes for isocitrate dehydrogenase (IDH), either IDH1 or IDH2 [3,4]. These IDH mutations are prevalent in lower-grade glioma and secondary glioblastoma [5]. In light of this knowledge, the classification based on histopathology has recently been amended and approved by the WHO [1]. Primary GBs, i.e. those wild-type for IDH, show prevalent chromosome 7 and 10 aneuploidy and frequent mutations in the promoter of the TERT gene coding for telomerase reverse transcriptase [6,7].
Results from -omics and next-generation genome sequencing of large numbers of GB consistently predicted their classification into three molecular subgroups: classical, proneural, and mesenchymal [[8], [9], [10], [11]]. The subclassification in IDH mutant versus IDH wild-type glioma described above has recently been supported and refined by genome-wide methylation profile analysis [12]. Thus, the methylomic profile has become the gold standard for molecular classification of gliomas and is now routinely used in neuropathology for clinical purposes. Besides identification of molecular diagnosis markers of gliomas, genomic studies have highlighted the recurrence of chromosomal alterations leading to amplification of receptor tyrosine kinase (RTK) genes, such as EGFR and PDGFR, or loss of PTEN and CDKN2A tumor suppressor genes encoding inhibitors of RTK signaling [7,[13], [14], [15], [16], [17]]. In fact, 88% of glioma biopsies carry genetic alterations in RTKs and their downstream pathways. This reflects the primordial role of RTKs and their downstream signaling pathways during gliomagenesis.
Upon binding of growth factors, RTKs homo- and/or heterodimerize, become autophosphorylated, activated, and are rapidly internalized by distinct endocytic sorting machineries for subsequent downregulation or recycling [18,19]. Endocytosis of activated RTK-ligand complexes has been reported to occur via two different internalization routes: clathrin-mediated or clathrin-independent endocytosis (CME or CIE, respectively) [[20], [21], [22], [23]]. CME proceeds through clathrin-coated pits and vesicles that are formed by the sequential recruitment of heterotetrameric AP-2 adaptor protein complexes comprising α, β2, μ2 and σ2 adaptins and of clathrin triskelia composed of three clathrin heavy chains (CHC17) with tightly associated clathrin light chains (CLCs) [24,25]. The efficient progression through clathrin-coated vesicle (CCV) initiation, formation, and scission is mediated by the orchestrated action of multiple accessory components, among them FCHo proteins, intersectins, and EPS15 to modulate recruited AP-2 [[26], [27], [28]], AP180/CALM, Dab2 and other alternative adaptors to expand cargo selection [29,30], Bin/Amphiphysin/Rvs (BAR) and Src Homology (SH3) domain-containing proteins such as amphiphysins and endophilins for sensing and generating membrane curvature [31], and the mechanochemical enzyme dynamin to finally pinch-off the nascent CCV from the plasma membrane [32,33]. Even though CME has been considered the major pathway for the uptake and downregulation of RTKs, evidence suggests that also CIE significantly contributes to internalization of activated receptor populations. Clathrin-independent uptake requires endophilin A1–3 as cargo adaptors and has thus been called fast endophilin-mediated endocytosis (FEME) [[34], [35], [36], [37]]. The consensus is that cells exposed to stimuli (e.g., growth factors) can internalize a significant fraction of activated RTKs via tubulo-vesicular carriers of the FEME pathway for efficient and fast RTK downregulation [20].
While the genetic basis of enhanced growth factor activity in GB has been established, regulatory mechanisms leading to strong RTK dependency in GB have remained elusive. To identify novel targets and regulatory mechanisms, we initiated an unbiased, deep and quantitative proteome analysis of glioma biopsies sampled from patients with various glioma grades and genetic subtypes. The major finding is that multiple endocytic machinery components involved in the formation of CME and/or FEME carriers are significantly reduced in patients irrespective of glioma grade and subclass. Building on our analyses, we here propose that impaired endocytosis provides a selective advantage for glioma tumor progression through increased growth factor sensing and prolonged RTK signaling from the cell surface.
2. Materials and methods
2.1. Patient biopsies
Biopsy samples were collected from patients after obtaining informed consent according to the guidelines of the local ethical committee (Ethikkommission Nordwestschweiz, #42/10). Samples were collected from patients undergoing surgical removal of gliomas. To match the glioma samples against control tissue, biopsies were collected from patients undergoing surgery for epilepsy, as already described in a number of previous studies [[38], [39], [40], [41], [42], [43], [44], [45], [46]]. Additional control biopsies from white matter were used for quality control by immunoblotting (Fig. S3). Immediately after resection, the tissue was snap-frozen and kept in liquid nitrogen until use.
2.2. Protein and peptide preparation, mass spectrometric and bioinformatic analysis
Biopsies ranging from 25 to 258 mg were minced with a razor blade and transferred into a 1.5 mL microtube containing 8 M urea in 50 mM Tris-HCl, pH 8.0, 75 mM NaCl (3 μL buffer per mg biopsy wet weight). The tissue was ground with a Kimble Pellet Pestle mixer (Sigma Aldrich) fitted with a 1.5 mL pestle two times for 30 s. The homogenate was pelleted at 12′000 ×g for 10 min and the supernatant was collected. Proteins were reduced with 10 mM DTT at 55 °C for 30 min and alkylated with 50 mM iodoacetamide at room temperature for 15 min. The extract was applied to a PD-10 desalting column (GE Healthcare) and proteins were eluted with 4 M urea in 50 mM Tris-HCl, pH 8.0, 75 mM NaCl. 1 mL fractions were collected and each fraction was measured at 280 nm and the protein-containing fractions were pooled.
Protein digestion was with two aliquots of endoproteinase LysC (1:100 enzyme to protein ratio each, Wako chemicals) for 2 h at 37 °C. The urea concentration was lowered to 2 M with 50 mM Tris-HCl, pH 8.0, 75 mM NaCl and digestion was continued overnight at 37 °C with trypsin (1:50, Sigma-Aldrich #T6567) followed by an additional trypsin aliquot (1,50) for two 2 h. Digestion was stopped with TFA to 1% final concentration. The peptides of the control and glioma extracts were desalted on SepPak cartridges (Sigma-Aldrich) with 0.1% TFA and bound peptides were eluted with 80% acetonitrile/0.1% TFA. The desalted peptide pools were dried in a SpeedVac.
To reduce peptide complexity, individual digests of the glioma biopsies were injected onto a Vydac 218TPN column (Dr. Maisch). Peptides were eluted at 30 μL/min from solvent A (20 mM ammonium formate, pH 4.5) to solvent B (80% acetonitrile containing 20 mM ammonium formate, pH 4.5) with the following gradient: 0 min 0% solvent B, 10 min 0% solvent B, 110 min 50% solvent B. 2-min fractions were collected into a 96-well microtiter plate. Following the HPLC run of the glioma digest, a second run under identical chromatographic conditions and equal peptide amount of pooled control was carried out. Fractions from the glioma run and the control run were individually pooled into six pools by combining every sixth fraction in the peptide-containing region of the chromatogram. The individual pools were dried and stored at −20 °C. For mass spectrometric analysis, the pools were dissolved in 30 μL 0.1% formic acid immediately before use.
The individual HPLC pools were analyzed by capillary liquid chromatography tandem MS (LC/MS/MS) using a separating column (0.075 mm × 30 cm) packed with Reprosil C18 reverse-phase material (2.4 μm particle size, Dr. Maisch). The column was connected to an Orbitrap Lumos Tribrid instrument (Thermo Scientific). The solvents used for peptide separation were 0.1% formic acid in water (solvent A) and 80% acetonitrile containing 0.1% formic acid in water (solvent B). 2 μL of the individual pool was injected with an Easy-nLC 1200 capillary pump (Thermo Scientific) set to 0.3 μL/min. A linear gradient from 0 to 35% solvent B in solvent A in 160 min was delivered at a flow rate of 0.25 μL/min. The eluting peptides were ionized at 2.5 kV. The mass spectrometer was operated in data-dependent mode. The precursor scan was done in the Orbitrap set to 120′000 resolution, while the fragment ions were mass analyzed in the LTQ instrument. The instrument was set to do as many fragmentations as possible within 3 s before returning to the full-scan mode and initiating another round of fragmentation. All fractions from each individual glioma biopsy and control sample were run in technical triplicates.
For protein identification, the MS/MS spectra were searched against the H. sapiens databank from SwissProt with Proteome Discoverer 2.2 (Thermo Scientific) set to Mascot and Sequest HT search engines with 10 ppm precursor ion tolerance. The fragment ions were set to 0.5 Da tolerance. The following modifications were used during the search: carbamidomethyl-cysteine as fixed modification, oxidized methionine, and protein N-terminal acetylation as variable modifications. The peptide search matches were set to 1% false discovery rate. For relative protein quantification, feature detection with the Minora module (Thermo Scientific, Reinach, Switzerland) was included into the search algorithm. The output of the Proteome Discoverer search was further analyzed with the Perseus software to find regulated proteins in the data set [47].
The generated proteomic glioma data inventory was uploaded to the public PRIDE database repository (project accession number: PXD014606, https://www.ebi.ac.uk/pride/archive/).
2.3. Primary cell culture and cell surface biotinylation assay
Patient-derived primary cells obtained from glioma biopsies (BTB152 and BTB251) were maintained and grown in Dulbecco's modified Eagle's medium/nutrient mixture F-12 (DMEM/F-12) (Sigma-Aldrich #D8062) supplemented with 10% fetal calf serum (FCS), 100 units/mL streptomycin and penicillin, and 30 mM HEPES at 37 °C in 5% CO2. Cells were kept in culture for maximally four passages.
To determine the relative fraction of the transferrin receptor (TfR) at the cell surface in steady-state, primary cells were cultured in 6-well clusters, starved with serum-free medium, incubated with 20 μg/mL biotin-labeled transferrin (Tf) (Sigma-Aldrich #T3915) in serum-free medium for 30 min at 4 °C, extensively washed with ice-cold PBS, harvested with lysis buffer (PBS containing 1% Triton X-100, 0.5% deoxycholate, 2 mM PMSF, and protease inhibitor cocktail), and the postnuclear supernatant (10′000 ×g for 10 min at 4 °C) was boiled in SDS sample buffer at 95 °C for 5 min.
To measure EGFR protein levels at the plasma membrane in steady-state, primary cells prepared in 6-well clusters were cell surface-biotinylated using 1 mg/mL sulfo-NHS-SS-biotin (ProteoChem #b2104) in PBS at 4 °C for 30 min. After incubating the cells with 50 mM glycine or NH4Cl in PBS for 5 min to quench free biotin and three additional washes, the cells were lysed as above and the postnuclear supernatant was collected. A fraction of the supernatant was used as loading control for total protein lysate. The postnuclear supernatant was incubated for 1 h at 4 °C with 20 μL streptavidin agarose beads (Thermo Fisher Scientific #20357). The beads were extensively washed with ice-cold PBS and boiled as above. Lysates or pull-downs were analyzed by SDS-gel electrophoresis and immunoblotting as described below.
2.4. Gel electrophoresis and immunoblot analysis
Proteins separated by SDS-gel electrophoresis (7.5–12.5% polyacrylamide) were transferred to Immobilon-PSQ PVDF membranes (Merck Millipore). After blocking with 5% non-fat dry milk or bovine serum albumin (BSA) in TBS (50 mM Tris·HCl, pH 7.6, 150 mM NaCl) with 0.1% Tween-20 (TBST) for 1 h, the membranes were probed with primary antibodies in 1% BSA in TBST for 2 h at room temperature or overnight at 4 °C, followed by incubation with HRP-coupled secondary antibodies in 1% BSA in TBST for 1 h at room temperature. Immobilon Western Chemiluminescent HRP Substrate (Merck Millipore) was used for detection, a Fusion Vilber Lourmat Imaging System for imaging, and Fiji software for quantitation.
For immunoblotting, mouse anti-α-adaptin (BD Biosciences #610501; 1:5′000), goat anti-α-adaptin (Everest Biotech #EB11875; 1:2′000), anti-β1/2-adaptin (BD Biosciences #610381; 1:5′000), mouse anti-actin (Merck Millipore #MAB1501; 1:100′000), rabbit anti-amphiphysin (Abcam #ab52646; 1:1′000), mouse anti-CHC17 (made from TD.1 hybridoma; 1:200), rabbit anti-Dab2 (Abcam #ab33441; 1:1′000), mouse anti-dynamin-1/2 (Merck Millipore #MABT188; 1:2′000), rabbit anti-EGFR (Cell Signaling Technology #4267; 1:2′000), rabbit anti-FCHo2 (Bethyl Laboratories #A304-560A; 1:1′000), mouse anti-HIP1R (BD Biosciences #612118; 1:1′000), rabbit pan-Akt (Cell Signaling Technology #4685; 1:2′000), rabbit phospho-Akt (Ser473) (Cell Signaling Technology #4060; 1:2′000), rabbit anti-EndoA1 (Bio-Rad #AHP2723; 1:5′000), mouse anti-AP180 (Sigma-Aldrich #A4825; 1:1′000), and mouse anti-TfR (Thermo Fisher Scientific #13–6890; 1:2′000) antibodies were used.
As secondary antibodies for immunoblotting, HRP-labeled goat anti-rabbit (Sigma-Aldrich #A0545; 1:10′000), goat anti-mouse (Sigma-Aldrich #A0168; 1:10′000), and rabbit anti-goat (Sigma-Aldrich #A8919; 1:5′000) immunoglobulin antibodies were used. To detect biotinylated proteins on blots, streptavidin-HRP (Thermo Fisher Scientific #434323; 1:10′000) was used.
2.5. Histology
Biopsy samples were formalin-fixed, paraffin-embedded, and stained with hematoxylin and eosin as performed previously (Bancroft, John D and Cook, H.C., Manual of histological techniques). Paraffin sections were photographed with an Olympus slide scanner (Münster, Germany).
3. Results
3.1. Proteomic analysis of glioma biopsies
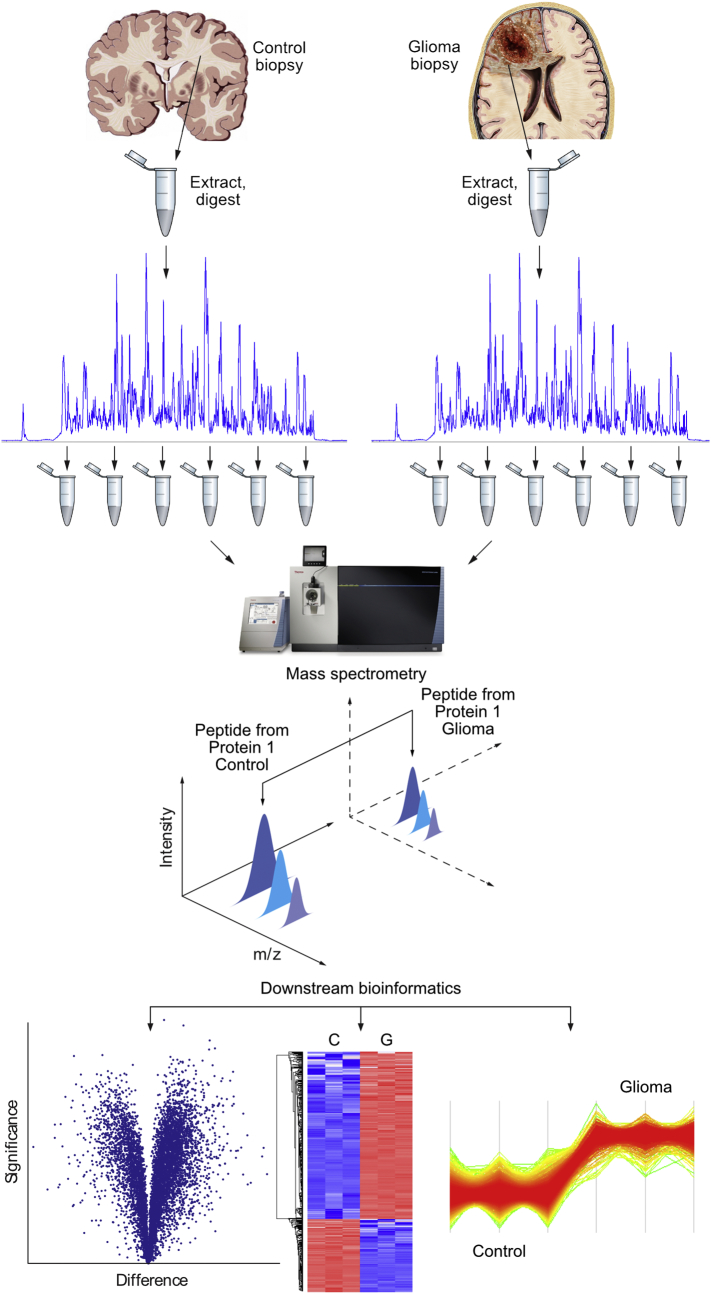
Initially, we performed proteomic profiling of eight biopsies from patients with grade II to IV gliomas. The mutational classification of the tumors, the clinical characteristics, tumor grading, details of the surgical resection, and overall survival data are summarized in Supplementary Tables S1 and S2. As control sample, white matter surgical specimen from five patients were extracted and pooled. Pooling of individual controls is a common technique in proteomic analysis and is used for levelling out individual variation to obtain a constant basis for statistical analysis [48]. The use of brain tissue from epileptic patients for control purposes is common and has been proven to be a reliable strategy for the comparison with tumor biopsy material [[38], [39], [40], [41], [42], [43], [44], [45], [46]]. In a similar manner to control, the glioma biopsies were extracted and matched in protein amount to the control sample. Glioma biopsies and controls were digested with endoproteinase LysC and trypsin (Fig. 1). To reduce peptide complexity, the digests were individually separated by reverse-phase chromatography and collected in six fractions. The individual fractions were then analyzed by high-resolution mass spectrometry. After peptide identification, the intensity for every protein occurring in both control and glioma biopsy was extracted and a library of differentially expressed proteins was established for further analysis [47].
Fig. 1.
Differential proteomics of glioma and control biopsies.
Proteins from control and glioma biopsies were extracted and digested with a combination of endoproteinase LysC and trypsin. The digests from control and glioma biopsies were separated individually by reverse-phase HPLC and pooled into six fractions. Each control and glioma-derived pool was separately analyzed by high-resolution mass spectrometry and the peptide intensities were compared and quantitated between the control and the respective glioma pool. From the peptide intensities the protein intensities were calculated and the data were statistically evaluated to find proteins either up- or downregulated in glioma. Proteins whose expression was significantly up- or downregulated were then functionally categorized by the DAVID software package and further processed with REVIGO into similar semantic classes.
3.2. Endocytic machinery components are downregulated in gliomas
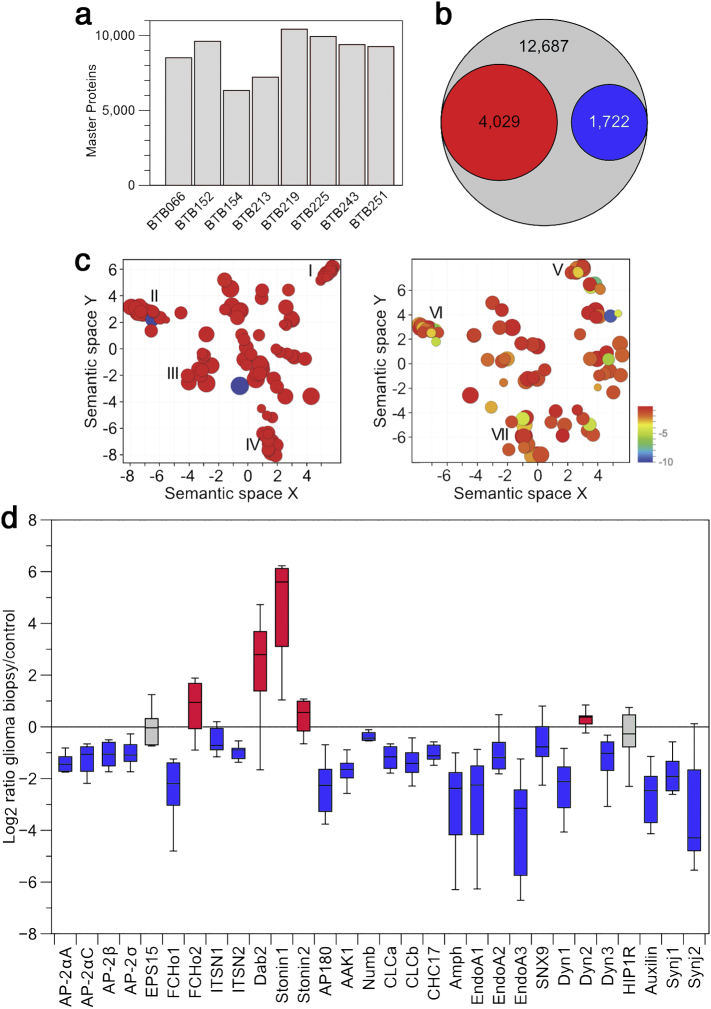
The proteomic inventory of each individual biopsy ranged from 6′331 to 10′430 proteins (Fig. 2a). By combining the identified proteins from all biopsies and filtering them for unique protein entries, a total number of 12′687 brain proteins were identified, from which 4′029 proteins were up- and 1′722 proteins downregulated by at least two-fold (Fig. 2b; for a compilation of all protein identifications from the individual biopsies, see also proteomics supplemental dataset S1A-H). Differentially expressed proteins were then summarized into functionally related classes by first submitting the dataset of proteins either two-fold up- or downregulated to the functional annotation tools of DAVID (https://david.ncifcrf.gov) [49]. Redundant gene ontology terms were removed and clusters of semantically similar gene ontology terms were obtained by REVIGO (http://revigo.irb.hr) [50] (Fig. 2c, for a compilation of all REVIGO terms, see also Table S3).
Fig. 2.
Quantitative proteomic analysis reveals a strong reduction of endocytic machinery components.
(a) Number of proteins identified from the individual glioma biopsies. (b) The number of proteins differentially upregulated (red) or downregulated (blue) by at least two-fold from the combined analysis of all glioma biopsies in comparison to the total of all proteins identified (gray). (c) Clustering of proteins by REVIGO that were upregulated (left) or downregulated (right) in glioma. The color code indicates log10 p-values according to the color bar. Gene ontology categories in Cluster I: IgG binding, β2-microglobulin binding, TAP binding, death receptor binding, Rho GTPase binding, platelet-derived growth factor binding, RNA polymerase II transcription factor binding; Cluster II: motor activity, histone deacetylase activity, cysteine/serine/threonine-type endopeptidase activity, ATP-dependent RNA helicase activity, chitinase activity, single-stranded DNA-dependent ATPase activity; Cluster III: guanine/thymine mispair binding, E-box binding, damaged DNA binding, mRNA 3′-UTR binding, translation elongation factor binding; Cluster IV: histone-lysine N-methyltransferase activity, histone acetyltransferase activity, histone kinase activity, protein kinase C activity, cyclin-dependent protein serine/threonine kinase activity; Cluster V: transmembrane transporter activity, voltage-gated sodium channel activity involved in cardiac muscle cell action potential, inorganic anion exchanger activity, hydrogen ion transmembrane transporter activity; cluster VI: neuropeptide hormone activity, neuroligin family protein binding, SH3 domain binding, clathrin binding; cluster VII: phosphatidylserine binding, calcium-dependent phospholipid binding, calcium ion binding, glycine binding. (d) Proteomic quantification of up- (red) and downregulated (blue) endocytic machinery components by plotting the log2 ratio of the individual endocytic components in glioma biopsies compared to control. Box plots show the median, the minima and maxima, the median and the first and third quartile, with whiskers representing the range. Full names of proteins can be found in Table S4. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
From the biological classification of downregulated proteins, we focused on the very tight semantic cluster VI that, among others, was annotated as “clathrin-binding cluster” (Fig. 2c). For the proteins present in this cluster, the log2-transformed ratio between the individual glioma biopsies and the control was calculated, which revealed a predominant reduction of endocytic machinery components involved in all stages of CCV and CIE carrier production at the plasma membrane [20,23], including initiators, main and alternative structural coat constituents, coat-associated kinases, membrane curvature sensors and benders, and factors involved in scission and uncoating of transport carriers (Fig. 2d, see also Table S4).
The key players in CCV production with well-characterized phenotypes are AP-2, clathrin, and dynamin. With the exception of dynamin 2, which is only marginally expressed in brain tissue compared to dynamin 1 and 3 [33], all these components showed at least a 2-fold reduction in protein amount. Furthermore, the proteomic quantification was very robust, with a standard deviation of 1–16% of the mean of the log2-transformed glioma-to-control biopsy ratios (Fig. S2a). The most strongly affected proteins in our analyses were the endophilins A1–3 (EndoA1–3). Individual isoforms even displayed a depletion by up to 100-fold (Fig. 2d). Even though EndoA1–3 have been characterized as part of the classical CME machinery, they are now recognized to also be involved in CIE pathways, in particular the FEME pathway [34,35]. Since also dynamin protein levels are reduced, on which CME and several CIE pathways depend, gliomas are likely to be globally impaired in endocytosis.
While the vast majority of endocytic machinery proteins was found to be robustly reduced, several alternative adaptors, including Dab2 and stonin 1 and 2, as well as the endocytic pioneer FCHo2 were conspicuously upregulated (Fig. 2d). Upregulation of Dab2 or the stonins might potentially provide an AP-2-independent rescue mechanism to compensate for the lack of uptake of LDLR or synaptic vesicle components, respectively [51,52]. Conversely, FCHo1 and 2 showed opposite regulation (Fig. 2d), potentially indicating different functionalities.
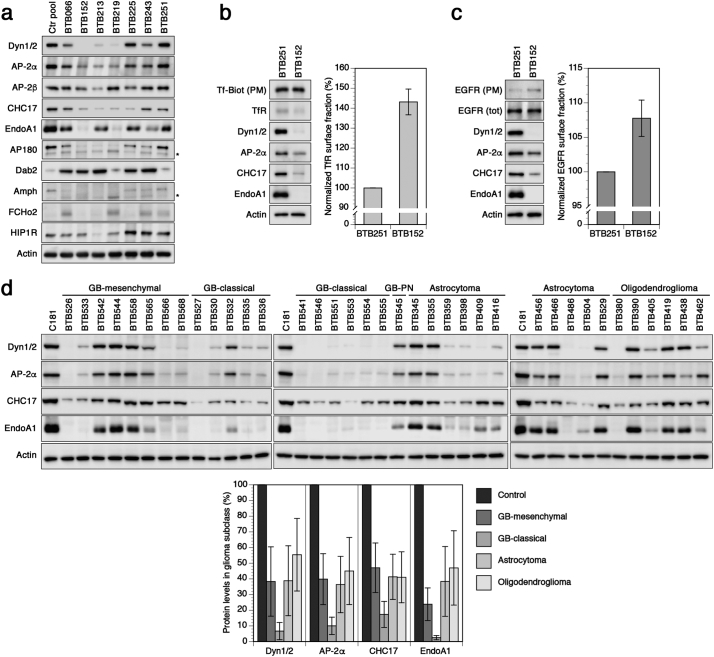
To validate the proteomic data, we analyzed a few representative components along the various stages of endocytosis by immunoblotting (Fig. 3a). First, we tested two subunits of the AP-2 complex, a core component in CME initiation and progression. Immunoblotting for the major, brain-specific α-adaptin (αA) [53] produced signals closely paralleling our proteomic data, with particularly low expression in BTB152, 213, and 219 (Fig. 3a). A reduction of β-subunits could also be observed. However, because the available antibody is not specific for β2 of AP-2, but also recognizes β1 of the AP-1 complex involved in intracellular trafficking at the TGN-to-endosome interface [54,55], the apparent effect is lower. The neuron-specific cargo adaptor AP180 was prominently depleted from most biopsies as well. In agreement with our proteomic data, immunoblots further demonstrated a robust reduction of the clathrin heavy chain (CHC17).
Fig. 3.
Downregulation of endocytic machinery components is a common trait of gliomas with different grades and subclasses.
(a) Immunoblot analysis of selected endocytic machinery components. Full names of proteins can be found in Table S4. To detect AP-2, we used an antibody specifically recognizing the brain-specific αA-subunit of AP-2. Asterisks indicate unspecific cross-reacting proteins. (b) Altered steady-state distribution of TfR between primary cells derived from BTB251 and BTB152 with mild and strong reduction of endocytic machinery, respectively. Accumulation of TfR at the cell surface in BTB152 is quantitated in percent relative to BTB251 (mean and standard deviation of four independent experiments). BTB-derived primary cells also displayed a reduction in endocytic machinery components (Dyn 1/2, AP-2 α, CHC17, EndoA1). (c) Altered steady-state distribution of EGFR. Normalized EGFR surface fraction is quantitated as in (b) (mean and standard deviation of four independent experiments). (d) Key endocytic machinery components (Dyn1/2, AP-2α, CHC17, EndoA1) are downregulated in various glioma subtypes, GB (classical), GB (mesenchymal), GB (PN), astrocytoma and oligodendroglioma. GB (classical), GB (mesenchymal), GB (PN) have been selected based on methylomic classifiers [12]; astrocytoma have been selected based on the IDH1/2 mutation status and absence of 1p19q codel; oligodendroglioma have been selected based on IDH1/2 mutation status and 1p19q codel [1]. Protein levels were quantified and plotted below in percent of the control sample of white matter mean and standard deviation of n = 8 GB (IDHwt, mesenchymal), n = 8 GB (IDHwt, classical), n = 11 astrocytoma (IDHmut), and n = 6 oligodendroglioma (IDHmut + 1p19q codel).
The scission of nascent carriers of CME and most CIE is driven by dynamin. Probing the glioma homogenates with an antibody against dynamin 1/2 revealed a strong downregulation, however, with only a modest decrease in BTB225 and BTB251 (Fig. 3a). EndoA1, which is also involved in the clathrin-independent FEME pathway, was severely depleted in all analyzed biopsies (Fig. 3a). Among the endophilins, EndoA1 is the most abundant and thus likely most relevant endophilin in brain. Previous studies characterizing the FEME pathway mainly relied on EndoA2 due to the use of non-neuronal HeLa and BSC1 cells [34,35]. It is interesting to note that downregulation of EndoA1 is associated with a number of cancers, such as breast carcinoma [56,57], laryngeal carcinoma [58], lung cancer [59], pituitary adenomas [60], and urothelial carcinoma [61].
Immunoblots of amphiphysin, HIP1R, FCHo2 and Dab2 closely mirrored our proteomics data and showed either downregulation, upregulation, or no regulation at all (Figs. 2d and 3a). In addition, using EGFR as the prototype RTK, we consistently found higher levels of the receptor protein in all biopsies compared to normal white matter, regardless of EGFR copy number, tumor histology, grade, or genetic background (Fig. S2b). Since the extent of machinery component downregulation varied among individual glioma biopsies, we raised the question how variable expression of these proteins is among control matter specimens. Indeed, while individual glioma biopsies were always robustly depleted compared to control (Figs. 2d, 3a and d), individual control white matter samples were fairly constant in endocytic protein levels and only differed ~3% (Fig. S3). In summary, the immunoblot analysis strongly corroborated our findings by quantitative proteomics and suggested a strong defect of endocytosis in glioma tumors.
3.3. Concerted endocytic machinery downregulation alters receptor distribution
Our proteomic results showed that most machinery components involved in CME and the FEME pathway are considerably downregulated in gliomas. Downregulation of these pathways is known to impede efficient receptor internalization [20,62,63], thereby shifting their steady-state distribution towards the cell surface. To test this experimentally, we compared the cell surface fraction of the transferrin receptor (TfR) and EGFR in two primary cell cultures derived from a biopsy with a comparatively moderate (BTB251) and from another one (BTB152) with a massive reduction of endocytic machinery components.
Immunoblots confirmed that the two primary cell lines retained the phenotypes of the respective biopsies with a pronounced depletion of AP-2, CHC17, EndoA1 and dynamin in BTB152 compared to BTB251 (Fig. 3b). To determine the fraction of TfR at the cell surface in steady-state, cultured primary cells were incubated with biotin-labeled transferrin (Tf-biotin) for 30 min at 4 °C, extensively washed, lysed and analyzed by immunoblot analysis. While both cell lines bound similar amounts of Tf-biotin, BTB152 showed ~30% reduction of total TfR and thus an increase of the surface transferrin receptor fraction of 43 ± 6%. Considering that BTB251 already displayed a partial loss of AP-2 and CHC17, this result is in agreement with previous reports showing a ~60–120% increase of the receptor at the cell surface upon inhibition or depletion of AP-2, clathrin, or dynamin in immortalized cell cultures [[64], [65], [66], [67], [68]].
In a similar manner, the plasma membrane population of EGFR was also determined in these cell lines using a cell surface biotinylation approach. Biotin-labeled EGFR was precipitated using streptavidin beads and then probed with an anti-EGFR antibody followed by immunoblot analysis. While the total amount of EGFR in both cell lines was indistinguishable, BTB152-derived primary cells had slightly higher steady-state levels of EGFR (8 ± 3%) at the plasma membrane than BTB251 cells, indicating a receptor shift towards the cell surface.
3.4. Defective endocytosis is a common trait among various glioma subclasses
To test whether downregulation of the endocytosis machinery is not limited to the eight glioma biopsies analyzed so far, but is also found among different glioma subclasses, we sampled additional 38 biopsies belonging to mesenchymal GB, classical GB, astrocytoma, and oligodendroglioma (see also Tables S5 and S6). Probing immunoblots of these biopsies with antibodies against dynamin, AP-2, CHC17 and EndoA1 demonstrated that these proteins were reduced in almost any instance, however, to different extents (Fig. 3d and e). The strongest depletion of endocytic machinery components was found in classical GB where all components were depleted to 10% or less of the control derived from non-tumorigenic white matter. Mesenchymal GB and anaplastic astrocytoma gliomas as well as oligodendroglioma equally displayed a reduction, however, with a more heterogeneous and less consistent pattern. Irrespective of any genetic background, all glioma subclasses have in common that they typically display elevated levels of phospho-Akt473 compared to non-tumor tissue (Fig. S2c). While endocytic machinery components among glioma subclasses were robustly and largely depleted, control white matter specimens showed a constant expression level throughout (Fig. S3).
To which extent defective endocytosis might contribute to increased RTK downstream signaling pathways is hard to be assessed from biopsy material. In line with previous findings [[69], [70], [71]], lacking RTK endocytosis results in increased activity of intracellular signaling pathways. Future studies with suitable model systems where multiple endocytic machinery components can be depleted simultaneously will allow to analyze the effect on distinct intracellular signaling pathways in the absence of additional changes found in tumors.
4. Discussion
In the present study, we have analyzed glioma biopsies of different grades by quantitative proteomics and uncovered a concerted reduction of multiple endocytic machinery components. By further expanding our analysis to other subclasses, we found that downregulation of endocytosis is a common trait of gliomas. Abnormal expression of individual proteins involved in endocytosis has been associated with human cancer for quite some time [72,73].
Endocytosis describes the uptake of external material or transmembrane cargo by membrane-enclosed transport vesicles or carriers. Depending on the characteristics and sorting determinants, cargo is internalized either by clathrin-dependent or -independent pathways (CME or CIE) [62,63]. A possible consequence of downregulation of proteins involved in endocytosis is impaired receptor internalization and hence increased steady-state levels on the surface and, upon growth factor activation, prolonged residence time at the plasma membrane leading to sustained RTK signaling and consequently to a selective growth advantage of tumor cells. The predominant machinery required for ligand-induced RTK endocytosis and downregulation is still debated, with reports showing the need for AP-2/clathrin or distinct CIE pathways [19,20,[74], [75], [76]]. An additional layer of complexity is introduced into this topic by the observation of a novel endophilin-mediated pathway (FEME) primed for the uptake of activated growth factor-bound RTKs [34,36]. A feature, however, that CME and some CIE pathways have in common is the mechanochemical enzyme dynamin to pinch off membrane-enclosed carriers. In our study, we found that dynamin, as well as machinery components of CME and CIE, including AP-2, clathrin, and endophilins, were simultaneously reduced in all mesenchymal glioblastomas tested and in many cases of all other glioma subclasses. Consistent with previous reports and our findings, depletion of endocytic machinery components impairs receptor internalization and thus accumulation at the cell surface. Impairment of endocytosis and hence prolonged growth factor signaling has been previously reported to modulate the activity of RTK downstream signaling pathways, including the AKT/PI3K, MAPK and PLC-γ pathway in cell lines [[69], [70], [71],[77], [78], [79]]. To which extent machinery component downregulation of dynamin-dependent CME and FEME perpetuates RTK autophosphorylation and thus activation of intracellular signaling cascades in gliomas remains to be further analyzed. Altered RTK distribution is most likely not only the result of CME downregulation, since depletion of AP-2 or clathrin alone was not reported to have a prominent effect on EGFR endocytosis [66]. More likely, this is also the effect of inhibition of the FEME pathway due to the reduction of EndoA1–3 (and the dynamins) (Figs. 2d and 3c), also in line with a recent publication by McMahon and colleagues where a triple knockdown of endophilin proteins prevented efficient RTK uptake in BSC1 and RPE1 cells [34].
Why are multiple components reduced and what is the mechanism responsible for concerted downregulation? Due to its fundamentality and essentiality, the endocytic pathway is most likely a very well buffered cellular system where depletion of single or a few endocytic network constituents does not result in severe phenotypes. The picture becomes more complex by the observation that, upon inactivation, CME and CIE crosstalk with each other and that dysfunction of either can boost compensatory traffic by the other [63,[80], [81], [82]]. Thus, downregulation of multiple proteins involved in CME and CIE might provide a strategy for glioma to efficiently inhibit RTK downregulation.
Given the fact that no mutation or any other genetic alterations could be reported in genes encoding downregulated endocytic machinery components in the analyzed gliomas, other mechanisms must account for the concerted modulation in reduced protein expression. One or more functional clusters of differentially expressed proteins that were upregulated among the analyzed glioma biopsy samples contained RNA/DNA-binding proteins (Fig. 2c and Table S3). Transcriptional repression by epigenetic regulation might provide a mechanism to simultaneously downregulate the expression of multiple genes. Current DNA methylomic analyses on glioma biopsies point towards expression control via gene methylation and transcriptional repression. The molecular and systematic characterization of these epigenetic reprogramming events go beyond the scope of the present report and will be the topic of future studies. Apart from increasing biopsy sample number, biopsy-free cell culture systems will be established to mimic effects of concerted machinery downregulation in vitro.
To the best of our knowledge, this is the first report showing a global and concerted reduction of multiple endocytic machinery components in cancer. In this regard, the endocytic machinery acts as previously suggested as tumor suppressor module inhibiting aberrant growth factor receptor signaling from the plasma membrane [83,84]. How CME, FEME and maybe other non-clathrin internalization pathways are regulated, individually or collectively, in gliomas for impairment of receptor endocytosis remains to be identified. Other factors commonly associated with RTK downregulation, such as the E3 ubiquitin protein ligase Cbl, did not show any clear tendency of regulation in our hands. Interestingly, a recently reported oncogenic factor, GOLM1, involved in modulating EGFR/RTK cell surface recycling in hepatocellular carcinoma (HCC) tissues, is upregulated in the biopsies [85] (proteomics supplemental datasets S1A-H). Thus, a vast repertoire of other strategic mechanisms exists to dictate RTKs' predominant presence at the cell surface apart from receptor amplification and endocytosis shutdown.
We thus think that reduction of endocytic machinery components represents one among other strategies to promote tumorgenesis via RTKs. In addition to revealing downregulation of endocytic components, the present study also provides an extensive inventory of proteins that are quantitatively regulated in glioma. This data repository as well as future data cohorts covering additional subtypes should be useful in the context of glioma research for finding and exploiting novel treatment strategies, but also for the understanding of other cancers and carcinogenesis in general.
Author contributions
P.J. and G.H. initiated the research project; D.P.B., J.-L.B., P.J., and G.H. designed research; D.P.B., M.-F.R., S.M., C.T., S.F. and P.J. performed research; D.P.B., S.M., C.T., S.F., P.J. analyzed data; and D.P.B., M.S., L.M., P.J., J.-L.B., and G.H. wrote the paper. D.P.B. submitted the manuscript and was handling the editorial process. All authors reviewed and revised the manuscript.
Declaration of Competing Interest
There are no conflicts of interest to disclose.
Acknowledgements
We very much thank the Hall laboratory for antibodies, and Cristina Prescianotto-Baschong, Faiza Noreen, and Primo Schär for preliminary experiments not included in this article. This work was supported by Grants 316030-164105 (P. Jenö), 31003A-162643 (M. Spiess) and PP00P3-176974 (G. Hutter) from the Swiss National Science Foundation. Further funding was received by the Department of Surgery from the University Hospital Basel.
Footnotes
The generated proteomic glioma data inventory was uploaded to the public PRIDE database repository (project accession number: PXD014606, https://www.ebi.ac.uk/pride/archive/). Supplementary data to this article can be found online at [https://doi.org/10.1016/j.ebiom.2019.07.039].
Contributor Information
Dominik P. Buser, Email: dominik-pascal.buser@unibas.ch.
Paul Jenö, Email: paul.jenoe@unibas.ch.
Jean-Louis Boulay, Email: jean-louis.boulay@unibas.ch.
Gregor Hutter, Email: gregor.hutter@usb.ch.
Appendix A. Supplementary data
Supplemental Materials (Fig. S1-3, Table S1-6).
Dataset S1A (BTB066).
Dataset S1B (BTB152).
Dataset S1C (BTB154).
Dataset S1D (BTB213).
Dataset S1E (BTB219).
Dataset S1F (BTB225).
Dataset S1G (BTB243).
Dataset S1H (BTB251).
References
- 1.Louis D.N., Perry A., Reifenberger G. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 2.Hegi M.E., Diserens A.C., Gorlia T. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352(10):997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 3.Parsons D.W., Jones S., Zhang X. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321(5897):1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Noushmehr H., Weisenberger D.J., Diefes K. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17(5):510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yan H., Parsons D.W., Jin G. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360(8):765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Killela P.J., Reitman Z.J., Jiao Y. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc Natl Acad Sci U S A. 2013;110(15):6021–6026. doi: 10.1073/pnas.1303607110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ozawa T., Riester M., Cheng Y.K. Most human non-GCIMP glioblastoma subtypes evolve from a common proneural-like precursor glioma. Cancer Cell. 2014;26(2):288–300. doi: 10.1016/j.ccr.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brennan C., Momota H., Hambardzumyan D. Glioblastoma subclasses can be defined by activity among signal transduction pathways and associated genomic alterations. PLoS One. 2009;4(11) doi: 10.1371/journal.pone.0007752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Phillips H.S., Kharbanda S., Chen R. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9(3):157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 10.Verhaak R.G., Hoadley K.A., Purdom E. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zacher A., Kaulich K., Stepanow S. Molecular diagnostics of gliomas using next generation sequencing of a Glioma-tailored gene panel. Brain Pathol. 2017;27(2):146–159. doi: 10.1111/bpa.12367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Capper D., Jones D.T.W., Sill M. DNA methylation-based classification of central nervous system tumours. Nature. 2018;555(7697):469–474. doi: 10.1038/nature26000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cancer Genome Atlas Research N Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Libermann T.A., Nusbaum H.R., Razon N. Amplification, enhanced expression and possible rearrangement of EGF receptor gene in primary human brain tumours of glial origin. Nature. 1985;313(5998):144–147. doi: 10.1038/313144a0. [DOI] [PubMed] [Google Scholar]
- 15.Muleris M., Almeida A., Dutrillaux A.M. Oncogene amplification in human gliomas: a molecular cytogenetic analysis. Oncogene. 1994;9(9):2717–2722. [PubMed] [Google Scholar]
- 16.Steck P.A., Pershouse M.A., Jasser S.A. Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat Genet. 1997;15(4):356–362. doi: 10.1038/ng0497-356. [DOI] [PubMed] [Google Scholar]
- 17.Sugawa N., Ekstrand A.J., James C.D., Collins V.P. Identical splicing of aberrant epidermal growth factor receptor transcripts from amplified rearranged genes in human glioblastomas. Proc Natl Acad Sci U S A. 1990;87(21):8602–8606. doi: 10.1073/pnas.87.21.8602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McMahon H.T., Boucrot E. Molecular mechanism and physiological functions of clathrin-mediated endocytosis. Nat Rev Mol Cell Biol. 2011;12(8):517–533. doi: 10.1038/nrm3151. [DOI] [PubMed] [Google Scholar]
- 19.Sorkin A., von Zastrow M. Endocytosis and signalling: intertwining molecular networks. Nat Rev Mol Cell Biol. 2009;10(9):609–622. doi: 10.1038/nrm2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferreira A.P.A., Boucrot E. Mechanisms of carrier formation during clathrin-independent endocytosis. Trends Cell Biol. 2018;28(3):188–200. doi: 10.1016/j.tcb.2017.11.004. [DOI] [PubMed] [Google Scholar]
- 21.Johannes L., Parton R.G., Bassereau P., Mayor S. Building endocytic pits without clathrin. Nat Rev Mol Cell Biol. 2015;16(5):311–321. doi: 10.1038/nrm3968. [DOI] [PubMed] [Google Scholar]
- 22.Kaksonen M., Roux A. Mechanisms of clathrin-mediated endocytosis. Nat Rev Mol Cell Biol. 2018;19(5):313–326. doi: 10.1038/nrm.2017.132. [DOI] [PubMed] [Google Scholar]
- 23.Mettlen M., Chen P.H., Srinivasan S., Danuser G., Schmid S.L. Regulation of clathrin-mediated endocytosis. Annu Rev Biochem. 2018;87:871–896. doi: 10.1146/annurev-biochem-062917-012644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cocucci E., Aguet F., Boulant S., Kirchhausen T. The first five seconds in the life of a clathrin-coated pit. Cell. 2012;150(3):495–507. doi: 10.1016/j.cell.2012.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kirchhausen T. Clathrin. Annu Rev Biochem. 2000;69:699–727. doi: 10.1146/annurev.biochem.69.1.699. [DOI] [PubMed] [Google Scholar]
- 26.Hollopeter G., Lange J.J., Zhang Y. The membrane-associated proteins FCHo and SGIP are allosteric activators of the AP2 clathrin adaptor complex. Elife. 2014;3 doi: 10.7554/eLife.03648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma L., Umasankar P.K., Wrobel A.G. Transient Fcho1/2Eps15/RAP-2 nanoclusters prime the AP-2 clathrin adaptor for cargo binding. Dev Cell. 2016;37(5):428–443. doi: 10.1016/j.devcel.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Umasankar P.K., Ma L., Thieman J.R. A clathrin coat assembly role for the muniscin protein central linker revealed by TALEN-mediated gene editing. Elife. 2014;3 doi: 10.7554/eLife.04137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bonifacino J.S., Traub L.M. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu Rev Biochem. 2003;72:395–447. doi: 10.1146/annurev.biochem.72.121801.161800. [DOI] [PubMed] [Google Scholar]
- 30.Mettlen M., Loerke D., Yarar D., Danuser G., Schmid S.L. Cargo- and adaptor-specific mechanisms regulate clathrin-mediated endocytosis. J Cell Biol. 2010;188(6):919–933. doi: 10.1083/jcb.200908078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haucke V., Kozlov M.M. Membrane remodeling in clathrin-mediated endocytosis. J Cell Sci. 2018;131(17) doi: 10.1242/jcs.216812. [DOI] [PubMed] [Google Scholar]
- 32.Antonny B., Burd C., De Camilli P. Membrane fission by dynamin: what we know and what we need to know. EMBO J. 2016;35(21):2270–2284. doi: 10.15252/embj.201694613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ferguson S.M., De Camilli P. Dynamin, a membrane-remodelling GTPase. Nat Rev Mol Cell Biol. 2012;13(2):75–88. doi: 10.1038/nrm3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boucrot E., Ferreira A.P., Almeida-Souza L. Endophilin marks and controls a clathrin-independent endocytic pathway. Nature. 2015;517(7535):460–465. doi: 10.1038/nature14067. [DOI] [PubMed] [Google Scholar]
- 35.Renard H.F., Simunovic M., Lemiere J. Endophilin-A2 functions in membrane scission in clathrin-independent endocytosis. Nature. 2015;517(7535):493–496. doi: 10.1038/nature14064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Watanabe S., Boucrot E. Fast and ultrafast endocytosis. Curr Opin Cell Biol. 2017;47:64–71. doi: 10.1016/j.ceb.2017.02.013. [DOI] [PubMed] [Google Scholar]
- 37.Chan Wah Hak L., Khan S., Di Meglio I. FBP17 and CIP4 recruit SHIP2 and lamellipodin to prime the plasma membrane for fast endophilin-mediated endocytosis. Nat Cell Biol. 2018;20(9):1023–1031. doi: 10.1038/s41556-018-0146-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heroux M.S., Chesnik M.A., Halligan B.D. Comprehensive characterization of glioblastoma tumor tissues for biomarker identification using mass spectrometry-based label-free quantitative proteomics. Physiol Genomics. 2014;46(13):467–481. doi: 10.1152/physiolgenomics.00034.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Collet B., Guitton N., Saikali S. Differential analysis of glioblastoma multiforme proteome by a 2D-DIGE approach. Proteome Sci. 2011;9(1):16. doi: 10.1186/1477-5956-9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lemee J.M., Com E., Clavreul A. Proteomic analysis of glioblastomas: what is the best brain control sample? J Proteomics. 2013;85:165–173. doi: 10.1016/j.jprot.2013.04.031. [DOI] [PubMed] [Google Scholar]
- 41.Arora A., Patil V., Kundu P. Serum biomarkers identification by iTRAQ and verification by MRM: S100A8/S100A9 levels predict tumor-stroma involvement and prognosis in Glioblastoma. Sci Rep. 2019;9(1):2749. doi: 10.1038/s41598-019-39067-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simeone P., Trerotola M., Urbanella A. A unique four-hub protein cluster associates to glioblastoma progression. PLoS One. 2014;9(7) doi: 10.1371/journal.pone.0103030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deighton R.F., McGregor R., Kemp J., McCulloch J., Whittle I.R. Glioma pathophysiology: insights emerging from proteomics. Brain Pathol. 2010;20(4):691–703. doi: 10.1111/j.1750-3639.2010.00376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Iwadate Y., Sakaida T., Hiwasa T. Molecular classification and survival prediction in human gliomas based on proteome analysis. Cancer Res. 2004;64(7):2496–2501. doi: 10.1158/0008-5472.can-03-1254. [DOI] [PubMed] [Google Scholar]
- 45.Chumbalkar V.C., Subhashini C., Dhople V.M. Differential protein expression in human gliomas and molecular insights. Proteomics. 2005;5(4):1167–1177. doi: 10.1002/pmic.200401202. [DOI] [PubMed] [Google Scholar]
- 46.Schwartz S.A., Weil R.J., Johnson M.D., Toms S.A., Caprioli R.M. Protein profiling in brain tumors using mass spectrometry: feasibility of a new technique for the analysis of protein expression. Clin Cancer Res. 2004;10(3):981–987. doi: 10.1158/1078-0432.ccr-0927-3. [DOI] [PubMed] [Google Scholar]
- 47.Tyanova S., Temu T., Sinitcyn P. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods. 2016;13(9):731–740. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- 48.Diz A.P., Truebano M., Skibinski D.O. The consequences of sample pooling in proteomics: an empirical study. Electrophoresis. 2009;30(17):2967–2975. doi: 10.1002/elps.200900210. [DOI] [PubMed] [Google Scholar]
- 49.da Huang W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 50.Supek F., Bosnjak M., Skunca N., Smuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One. 2011;6(7) doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maurer M.E., Cooper J.A. The adaptor protein Dab2 sorts LDL receptors into coated pits independently of AP-2 and ARH. J Cell Sci. 2006;119(Pt 20):4235–4246. doi: 10.1242/jcs.03217. [DOI] [PubMed] [Google Scholar]
- 52.Willox A.K., Royle S.J. Stonin 2 is a major adaptor protein for clathrin-mediated synaptic vesicle retrieval. Curr Biol. 2012;22(15):1435–1439. doi: 10.1016/j.cub.2012.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ball C.L., Hunt S.P., Robinson M.S. Expression and localization of alpha-adaptin isoforms. J Cell Sci. 1995;108(Pt 8):2865–2875. doi: 10.1242/jcs.108.8.2865. [DOI] [PubMed] [Google Scholar]
- 54.Guo Y., Sirkis D.W., Schekman R. Protein sorting at the trans-Golgi network. Annu Rev Cell Dev Biol. 2014;30:169–206. doi: 10.1146/annurev-cellbio-100913-013012. [DOI] [PubMed] [Google Scholar]
- 55.Robinson M.S. Adaptable adaptors for coated vesicles. Trends Cell Biol. 2004;14(4):167–174. doi: 10.1016/j.tcb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 56.Kannan A., Wells R.B., Sivakumar S. Mitochondrial reprogramming regulates breast cancer progression. Clin Cancer Res. 2016;22(13):3348–3360. doi: 10.1158/1078-0432.CCR-15-2456. [DOI] [PubMed] [Google Scholar]
- 57.Sinha S., Chunder N., Mukherjee N. Frequent deletion and methylation in SH3GL2 and CDKN2A loci are associated with early- and late-onset breast carcinoma. Ann Surg Oncol. 2008;15(4):1070–1080. doi: 10.1245/s10434-007-9790-0. [DOI] [PubMed] [Google Scholar]
- 58.Shang C., Fu W.N., Guo Y., Huang D.F., Sun K.L. Study of the SH3-domain GRB2-like 2 gene expression in laryngeal carcinoma. Chin Med J (Engl) 2007;120(5):385–388. [PubMed] [Google Scholar]
- 59.Dasgupta S., Jang J.S., Shao C. SH3GL2 is frequently deleted in non-small cell lung cancer and downregulates tumor growth by modulating EGFR signaling. J Mol Med (Berl) 2013;91(3):381–393. doi: 10.1007/s00109-012-0955-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Farrell W.E., Simpson D.J., Bicknell J.E., Talbot A.J., Bates A.S., Clayton R.N. Chromosome 9p deletions in invasive and noninvasive nonfunctional pituitary adenomas: the deleted region involves markers outside of the MTS1 and MTS2 genes. Cancer Res. 1997;57(13):2703–2709. [PubMed] [Google Scholar]
- 61.Majumdar S., Gong E.M., Di Vizio D. Loss of Sh3gl2/endophilin A1 is a common event in urothelial carcinoma that promotes malignant behavior. Neoplasia. 2013;15(7):749–760. doi: 10.1593/neo.121956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kirchhausen T., Owen D., Harrison S.C. Molecular structure, function, and dynamics of clathrin-mediated membrane traffic. Cold Spring Harb Perspect Biol. 2014;6(5):a016725. doi: 10.1101/cshperspect.a016725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mayor S., Parton R.G., Donaldson J.G. Clathrin-independent pathways of endocytosis. Cold Spring Harb Perspect Biol. 2014;6(6) doi: 10.1101/cshperspect.a016758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen J., Wang J., Meyers K.R., Enns C.A. Transferrin-directed internalization and cycling of transferrin receptor 2. Traffic. 2009;10(10):1488–1501. doi: 10.1111/j.1600-0854.2009.00961.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Damke H., Baba T., van der Bliek A.M., Schmid S.L. Clathrin-independent pinocytosis is induced in cells overexpressing a temperature-sensitive mutant of dynamin. J Cell Biol. 1995;131(1):69–80. doi: 10.1083/jcb.131.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hinrichsen L., Harborth J., Andrees L., Weber K., Ungewickell E.J. Effect of clathrin heavy chain- and alpha-adaptin-specific small inhibitory RNAs on endocytic accessory proteins and receptor trafficking in HeLa cells. J Biol Chem. 2003;278(46):45160–45170. doi: 10.1074/jbc.M307290200. [DOI] [PubMed] [Google Scholar]
- 67.Moskowitz H.S., Heuser J., McGraw T.E., Ryan T.A. Targeted chemical disruption of clathrin function in living cells. Mol Biol Cell. 2003;14(11):4437–4447. doi: 10.1091/mbc.E03-04-0230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Motley A., Bright N.A., Seaman M.N., Robinson M.S. Clathrin-mediated endocytosis in AP-2-depleted cells. J Cell Biol. 2003;162(5):909–918. doi: 10.1083/jcb.200305145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Brankatschk B., Wichert S.P., Johnson S.D., Schaad O., Rossner M.J., Gruenberg J. Regulation of the EGF transcriptional response by endocytic sorting. Sci Signal. 2012;5(215) doi: 10.1126/scisignal.2002351. ra21. [DOI] [PubMed] [Google Scholar]
- 70.Sousa L.P., Lax I., Shen H., Ferguson S.M., De Camilli P., Schlessinger J. Suppression of EGFR endocytosis by dynamin depletion reveals that EGFR signaling occurs primarily at the plasma membrane. Proc Natl Acad Sci U S A. 2012;109(12):4419–4424. doi: 10.1073/pnas.1200164109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vieira A.V., Lamaze C., Schmid S.L. Control of EGF receptor signaling by clathrin-mediated endocytosis. Science. 1996;274(5295):2086–2089. doi: 10.1126/science.274.5295.2086. [DOI] [PubMed] [Google Scholar]
- 72.Floyd S., De Camilli P. Endocytosis proteins and cancer: a potential link? Trends Cell Biol. 1998;8(8):299–301. doi: 10.1016/s0962-8924(98)01316-6. [DOI] [PubMed] [Google Scholar]
- 73.Mosesson Y., Mills G.B., Yarden Y. Derailed endocytosis: an emerging feature of cancer. Nat Rev Cancer. 2008;8(11):835–850. doi: 10.1038/nrc2521. [DOI] [PubMed] [Google Scholar]
- 74.Goh L.K., Sorkin A. Endocytosis of receptor tyrosine kinases. Cold Spring Harb Perspect Biol. 2013;5(5):a017459. doi: 10.1101/cshperspect.a017459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sigismund S., Avanzato D., Lanzetti L. Emerging functions of the EGFR in cancer. Mol Oncol. 2018;12(1):3–20. doi: 10.1002/1878-0261.12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tomas A., Futter C.E., Eden E.R. EGF receptor trafficking: consequences for signaling and cancer. Trends Cell Biol. 2014;24(1):26–34. doi: 10.1016/j.tcb.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Galperin E., Sorkin A. Endosomal targeting of MEK2 requires RAF, MEK kinase activity and clathrin-dependent endocytosis. Traffic. 2008;9(10):1776–1790. doi: 10.1111/j.1600-0854.2008.00788.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lill N.L., Sever N.I. Where EGF receptors transmit their signals. Sci Signal. 2012;5(243):pe41. doi: 10.1126/scisignal.2003341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Miaczynska M. Effects of membrane trafficking on signaling by receptor tyrosine kinases. Cold Spring Harb Perspect Biol. 2013;5(11):a009035. doi: 10.1101/cshperspect.a009035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Balklava Z., Pant S., Fares H., Grant B.D. Genome-wide analysis identifies a general requirement for polarity proteins in endocytic traffic. Nat Cell Biol. 2007;9(9):1066–1073. doi: 10.1038/ncb1627. [DOI] [PubMed] [Google Scholar]
- 81.Donaldson J.G., Johnson D.L., Dutta D. Rab and Arf G proteins in endosomal trafficking and cell surface homeostasis. Small GTPases. 2016;7(4):247–251. doi: 10.1080/21541248.2016.1212687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dutta D., Donaldson J.G. Sorting of clathrin-independent cargo proteins depends on Rab35 delivered by clathrin-mediated endocytosis. Traffic. 2015;16(9):994–1009. doi: 10.1111/tra.12302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schmid S.L. Reciprocal regulation of signaling and endocytosis: implications for the evolving cancer cell. J Cell Biol. 2017;216(9):2623–2632. doi: 10.1083/jcb.201705017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lanzetti L., Di Fiore P.P. Endocytosis and cancer: an 'insider' network with dangerous liaisons. Traffic. 2008;9(12):2011–2021. doi: 10.1111/j.1600-0854.2008.00816.x. [DOI] [PubMed] [Google Scholar]
- 85.Ye Q.H., Zhu W.W., Zhang J.B. GOLM1 modulates EGFR/RTK cell-surface recycling to drive hepatocellular carcinoma metastasis. Cancer Cell. 2016;30(3):444–458. doi: 10.1016/j.ccell.2016.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Materials (Fig. S1-3, Table S1-6).
Dataset S1A (BTB066).
Dataset S1B (BTB152).
Dataset S1C (BTB154).
Dataset S1D (BTB213).
Dataset S1E (BTB219).
Dataset S1F (BTB225).
Dataset S1G (BTB243).
Dataset S1H (BTB251).