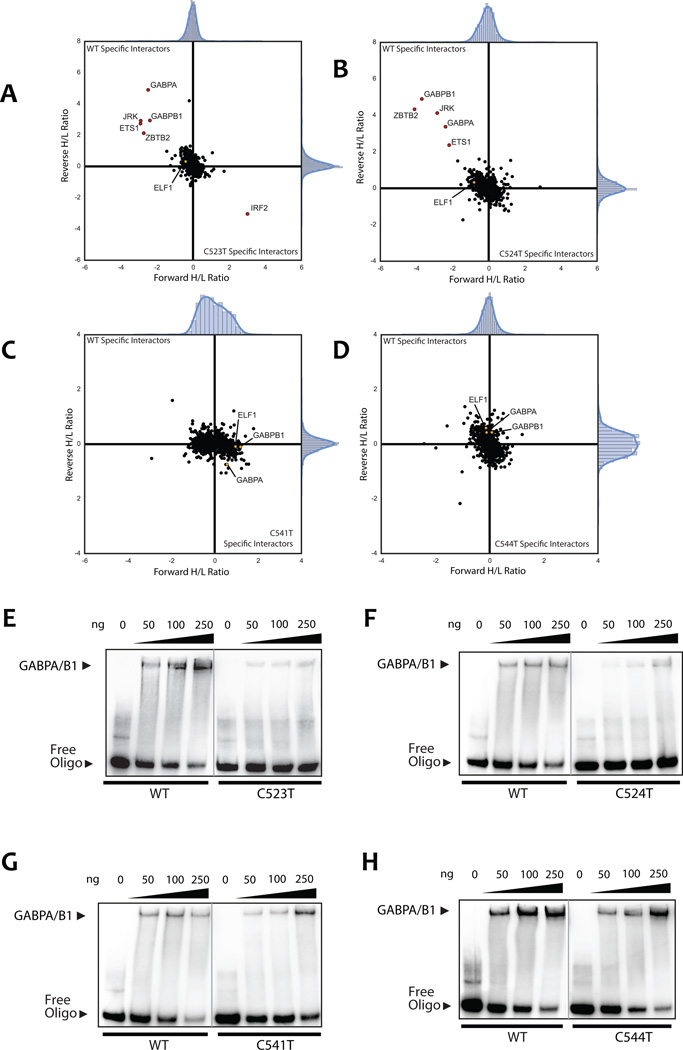
Figure 5.
AP-MS/MS identifies allele-specific protein-DNA interactions for hotspot SDHD promoter mutations. Custom oligos were 5’-biotinylated and designed to cover all analyzed mutation sites in the SDHD promoter as indicated in (Supplemental Table S3). Each mutation site was analyzed by two independent label-swapped experiments. ELF1, GABPA, and GABPB1, if not observed as significant interactors, are colored in yellow in the background cloud and noted by name. A, AP-MS/MS analysis of the C523T SDHD promoter mutation interactors. Interactors with a ratio of at least 3 IQR (inter-quartile range) in both replicate experiments are colored in red. Ratios are shown after log2 transformation. Labels were swapped between replicates to avoid labeling bias, hence specific interactors show a high ratio in one experiment and a low ratio in the other. B, AP-MS/MS analysis of the C524T SDHD promoter mutation interactors. C, AP-MS/MS analysis of the C541T SDHD promoter mutation interactors. D, AP-MS/MS analysis of the C544T SDHD promoter mutation interactors. E–H, band-shift experiments confirm SDHD WT-specific GABP binding at C523T and C524T mutation sites, but not C541T or C544T. Oligos were shifted using recombinant human protein as described in the materials and methods. E, band-shift analysis with the C523T SDHD promoter mutation oligo and recombinant human GABP (GABPA and GABPB1) protein. Band-shift experiments for each protein-oligo combination were resolved on the same gel at the same exposure. The grey line indicates where a single lane was cropped out for clarity. F–H, band-shift analysis with (F) C524T, (G) C541T, and (H) C544T SDHD promoter mutation oligonucleotides and recombinant human GABP protein.