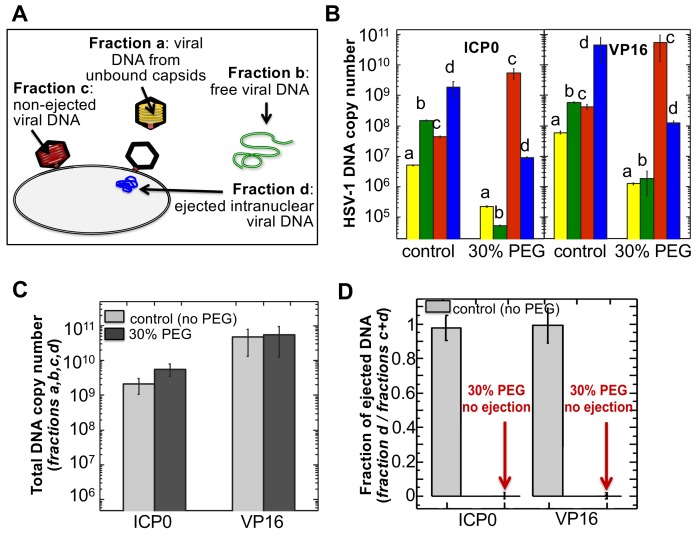
Figure 5. The amounts of HSV-1 DNA released into the nucleoplasm from capsids bound to NPCs quantified by qPCR after extraction from each fraction (a,b,c, and d).
Specific HSV-1 primers for VP16/UL48 and ICP0 genes were used as amplicons for the qPCR assays of viral DNA copy numbers.(A). Schematic of viral DNA fractions separated from the reconstituted capsid-nuclei system with pull-down assay. (B) Histograms show the DNA copy number for each of the four viral DNA-containing fractions. (C) Histograms show the total DNA copy number from fractions a, b, c, and d in the reconstituted capsid-nuclei system. (D) Fraction of DNA ejected from nuclei-bound capsids (fraction d/fractions c+d). After nuclei incubation with HSV-1 C-capsids at 37°C for 40 min, without osmolyte addition,~98% of all nuclei bound capsids ejected their DNA into nuclei. When the capsid pressure is turned off at 18 atm of external osmotic pressure (generated by addition of 30% w/w PEG 8 kDa), the ejection of DNA from capsids bound to nuclei is completely suppressed (fraction d/fractions c+d ~ 0.2%). qPCR DNA copy number quantification is based on a standard curve generated by serial dilution of a wild-type HSV-1 DNA with known DNA copy number. ICP0 and VP16 HSV-1 genes were quantified using specific primers. Error bars in B are standard deviations in DNA copy numbers from three independent qPCR reactions repeated at the same conditions. Error bars in C and D are progressed standard deviations.