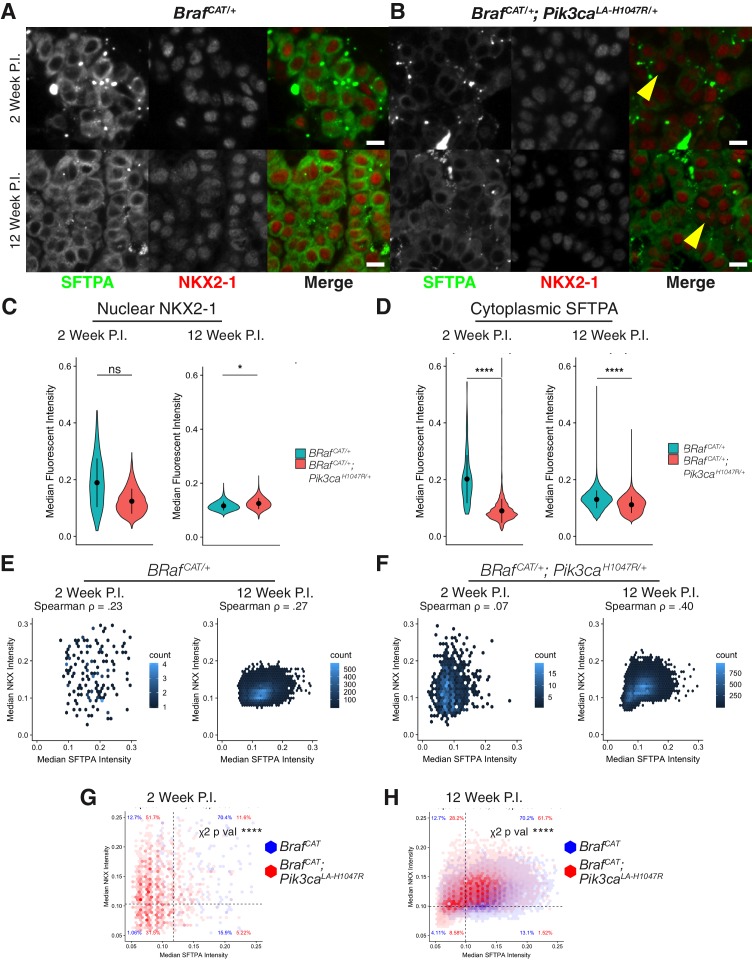
Figure 4. Expression levels and localization of lung lineage survival transcription factors are maintained in BRAFV600E/PI3KαH1047R driven tumors, Including those cells which have lost expression of markers of AT2 identity.
(A) BRAFV600E driven hyperplasia and tumors display widespread expression of both SFTPA and nuclear localization of the lung lineage transcription factor, NKX2-1, at 2 and 12 weeks post initiation. Scale bar = 10 um. (B) BRAFV600E/PI3KαH1047R driven hyperplasia and tumors show decreased SFTPA expression at 2 and 12 weeks post initiation. These tumors maintain broad nuclear expression of NKX2-1, including those cells with decreased SFTPA expression (yellow arrowheads). (C) Quantitation showing no significant difference in NKX2-1 immunoreactivity at 2 weeks post initiation, but a slight increase in nuclear NKX2-1 at 12 weeks post initiation. Wilcoxon rank sum p values = 0.2,. 02 respectively. (D) Significant reduction of SFTPA immunoreactivity seen in BRAFV600E/PI3KαH1047R driven hyperplasia and tumors at both 2 and 12 weeks post initiation. Wilcoxon rank sum p values = 5e-5, 4e-5 respectively. (E) Cytoplasmic SFTPA immunoreactivity plotted versus nuclear NKX2-1 immunoreactivity in BRAFV600E driven hyperplasia and tumors at 2 and 12 weeks post initiation. Similar association seen at both time points (Rho = 0.23,. 27 respectively). (F) Cytoplasmic SFTPA immunoreactivity plotted versus nuclear NKX2-1 immunoreactivity in BRAFV600E/PI3KαH1047R driven hyperplasia and tumors at 2 and 12 weeks post initiation. Relatively lower association seen at 2 weeks compared to 12 weeks (Rho = 0.07,. 40 respectively). (G) Overlay of BRAFV600E/PI3KαH1047R and BRAFV600E driven hyperplasia 2 weeks post initiation. Dashed line for each marker drawn at mean - one standard deviation of BRAFV600E driven tumors. BRAFV600E/PI3KαH1047R driven tumors show fewer SFTPA+, NKX2−1 + cells, most strongly accounted for by an increase in SFTPA-, NKX2−1 + cells. Chi square test associates genotype with distribution, p val <1e-5. (H) Overlay of BRAFV600E/PI3KαH1047R and BRAFV600E driven tumors 12 weeks post initiation. Dashed line for each marker drawn at mean - one standard deviation of BRAFV600E driven tumors. BRAFV600E/PI3KαH1047R driven tumors show fewer SFTPA+, NKX2−1 + cells, most strongly accounted for by an increase in SFTPA-, NKX2−1 + cells. Chi square test associates genotype with distribution, p val <1e-5.