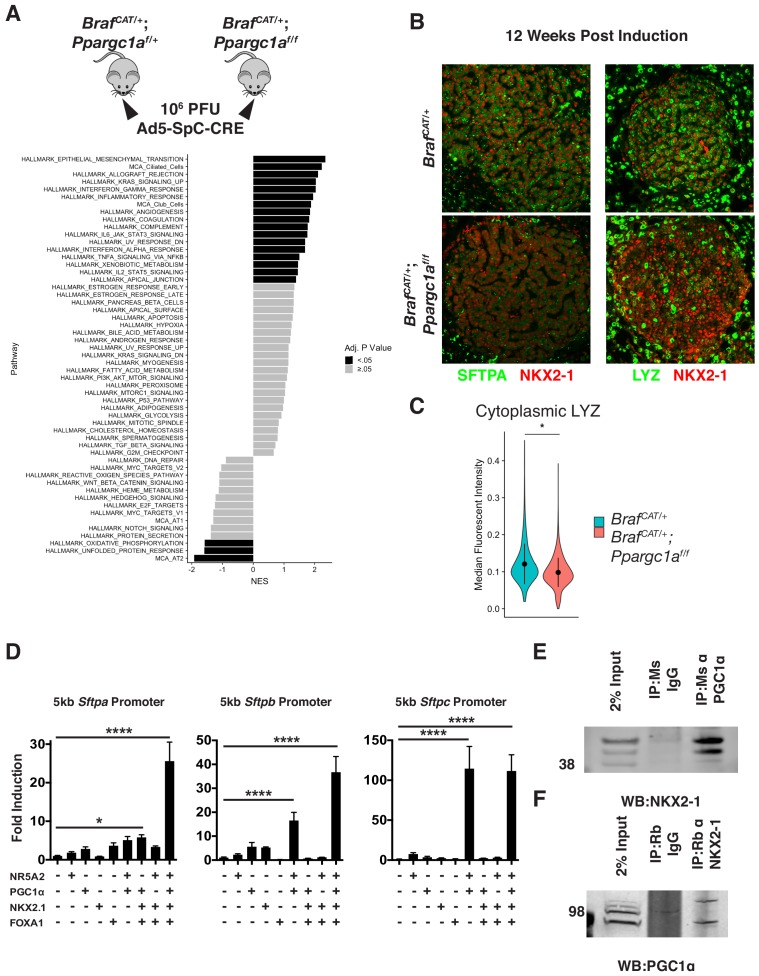
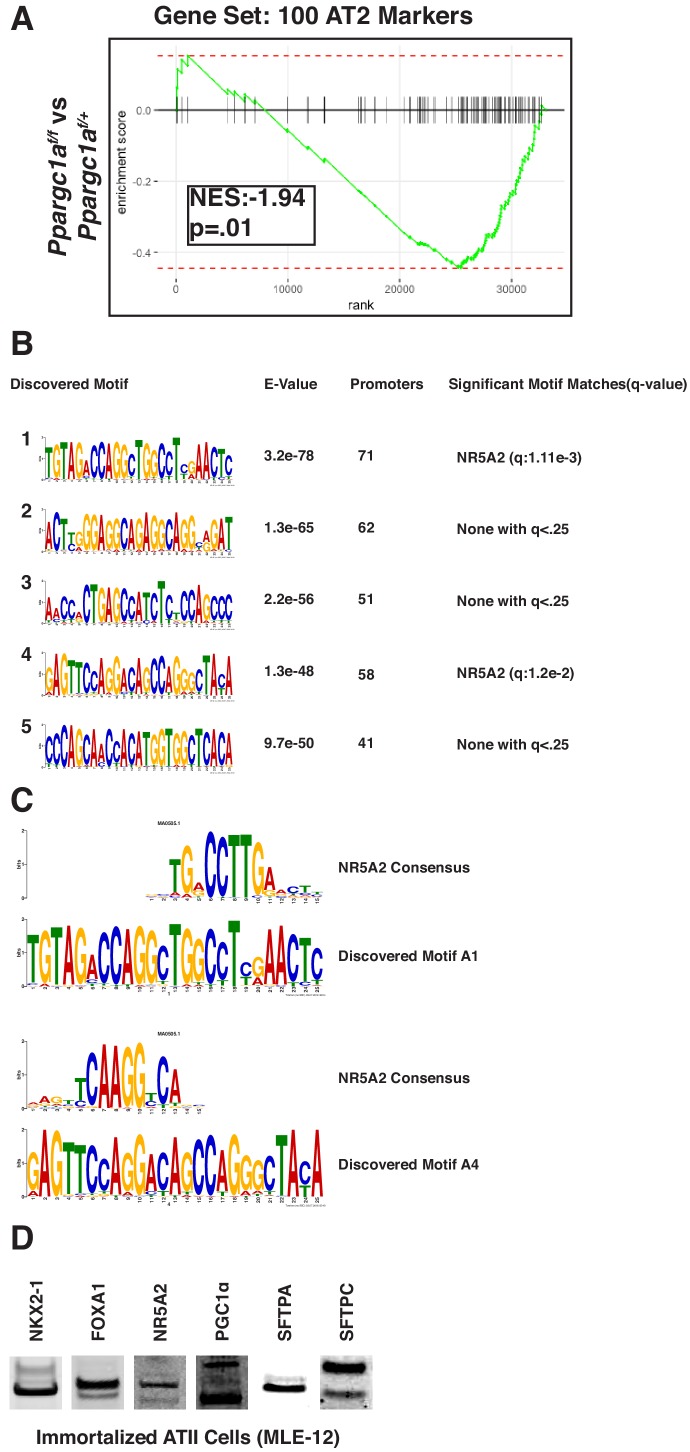
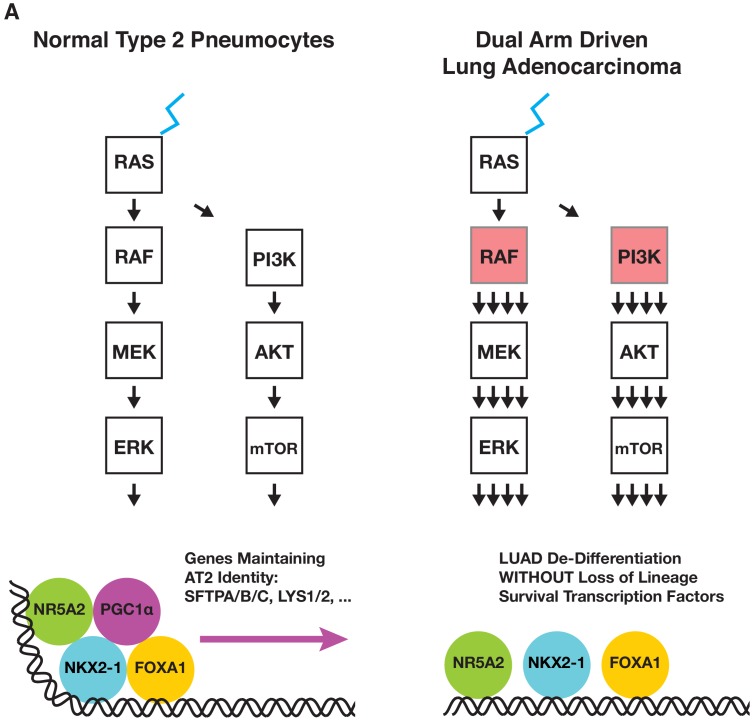
Figure 7. PGC1α is required for maintenance of lung identity in BRAFV600E driven tumors.
(A) Tumors were induced in cohorts of BrafCAT/+ and BrafCAT/+;Ppargc1af/f mice via intranasal instillation of 106 PFU Ad5-SpC-CRE and harvested from each genotype 12 weeks post tumor induction via tissue dissociation and FACS. GSEA analyses of hallmark pathways and lung identity gene sets. Black bars indicate adjP <.05, gray bars indicate Benjamini-Hochberg corrected enrichment statistic adjP ≥. 05. (B) Immunostaining confirms decreased expression of the AT2 markers SFTPA and LYZ in BRAFV600E/PGC1αNULL tumors. (C) Quantitation demonstrating a significant decrease of LYZ immunoreactivity in BRAFV600E/PGC1αNULL tumors. Wilcoxon rank sum p val. = 0.0288. (D) Luciferase assays in HEK293T cells demonstrating the cooperation of NKX2-1, FOXA1, PGC1α, and NR5A2 in transactivation of surfactant promoters. All three promoters showed significant induction by ordinary one-way ANOVA (p<0.0001). Comparison of individual groups to mock transfected controls by Dunnett’s test for multiple comparisons: (*) p=0.0189, (****) p<0.0001. (E) Co-Immunoprecipitation of NKX2-1 by immunoprecipitation with a mouse monoclonal antibody recognizing PGC1α but not with IgG. (F) Co-Immunoprecipitation of PGC1α by immunoprecipitation with a mouse monoclonal antibody recognizing NKX2-1 but not with mouse IgG.