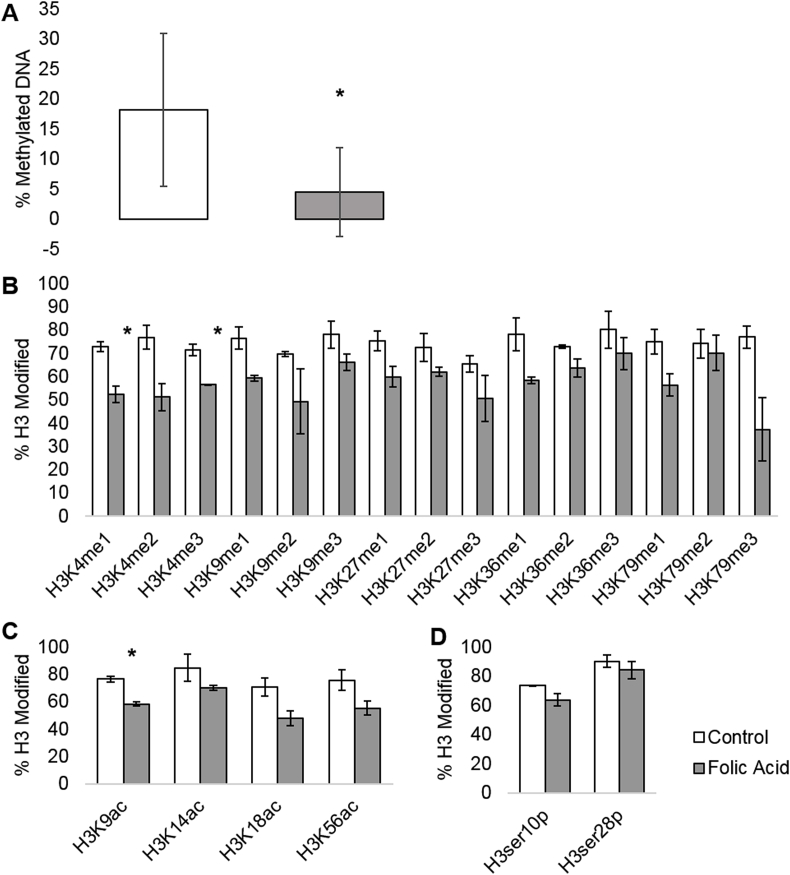
Fig. 3.
A. DNA methylation in SHSY5Y cells as determined by 5-mc ELISA assay; p = 0.048. Three replicates per treatment group were tested. Error bars are ± 1 standard deviation. B. Percentage of histone 3 (H3) modified with a particular methylation mark; p = 0.034 for H3K4me1, p = 0.073 for H3K4me2, p = 0.027 for H3K4me3, p = 0.065 for H3K9me1, p = 0.21 for H3K9me2, p = 0.18 for H3K9me3, p = 0.11 for H3K27me1, p = 0.18 for H3K27me2, p = 0.21 for H3K27me3, p = 0.094 for H3K36me1, p = 0.12 for H3K36me2, p = 0.28 for H3K36me3, p = 0.10 for H3K79me1, p = 0.40 for H3K79me2, and p = 0.10 for H3K79me3. Three replicates per treatment group were tested. Error bars are ± 1 standard deviation. C. Percentage of H3 modified with a particular acetylation mark; p = 0.018 for H3K9Ac, p = 0.21 for H3K14Ac, p = 0.10 for H3K18Ac, and p = 0.13 for H3K56Ac. Three replicates per treatment group were tested. Error bars are ± 1 standard deviation. D. Percentage of H3 modified with a particular phosphorylation mark; p = 0.13 for H3ser10p, and p = 0.31 for H3ser28p. Three replicates per treatment group were tested. Error bars are ± 1 standard deviation. An asterisk (*) indicates p < 0.05 using a paired two-tailed t-test.