Fig. 2.
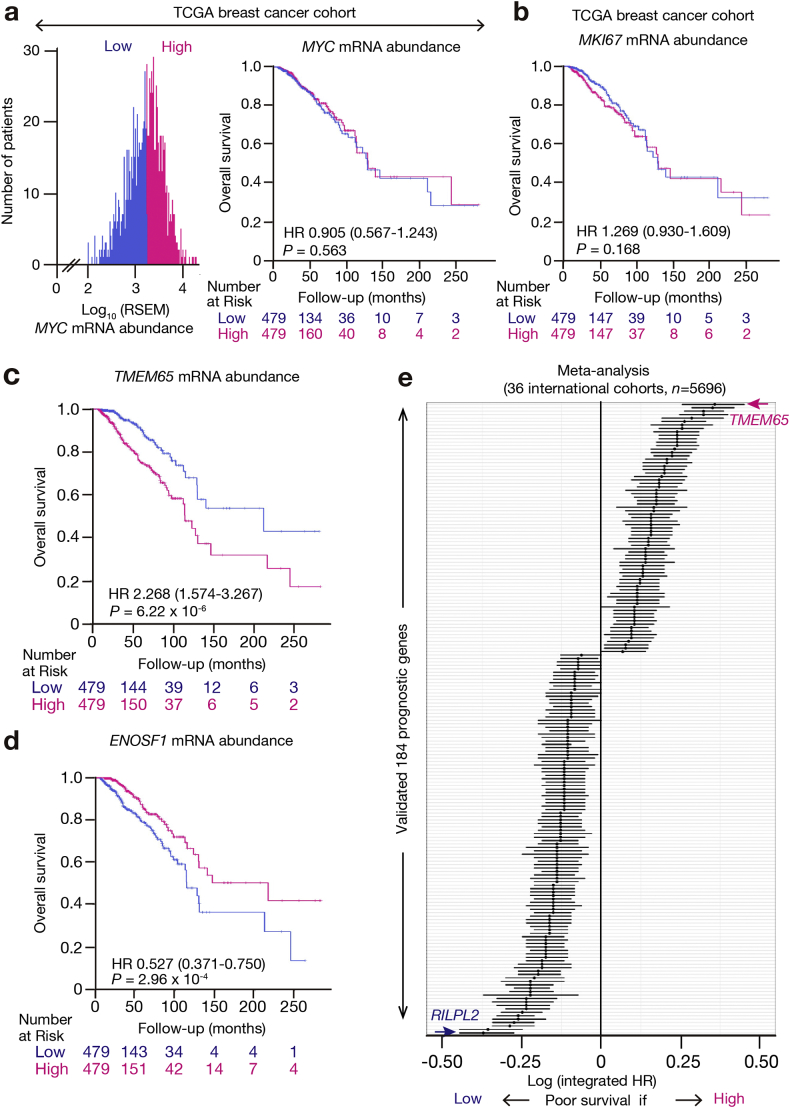
Identification of all prognosis-related genes in the TCGA breast cancer cohort and validation in 36 independent cohorts. (a) Distribution of MYC expression level (RSEM) among patients in the TCGA breast cancer cohort (left), and Kaplan-Meier curves of OS for these patients based on a MYC expression level higher or lower than the median (right). The HR, its 95% CI, and the log-rank P value are shown. (b) Kaplan-Meier curves of OS for the TCGA cohort based on MKI67 expression level. (c and d) Kaplan-Meier curves of OS for the TCGA cohort based on TMEM65 (c) and ENOSF1 (d) expression levels, respectively. The complete list of OS-related genes in this TCGA discovery cohort is provided in Supplementary Table S2. (e) Logarithm of the integrated HR for all 184 prognosis-related genes in the validation data sets. The complete list of these genes identified by meta-analysis is provided in Supplementary Table S3.
Identification of all prognosis-related genes in the TCGA breast cancer cohort and validation in 36 independent cohorts. (a) Distribution of MYC expression level (RSEM) among patients in the TCGA breast cancer cohort (left), and Kaplan-Meier curves of OS for these patients based on a MYC expression level higher or lower than the median (right). The HR, its 95% CI, and the log-rank P value are shown. (b) Kaplan-Meier curves of OS for the TCGA cohort based on MKI67 expression level. (c and d) Kaplan-Meier curves of OS for the TCGA cohort based on TMEM65 (c) and ENOSF1 (d) expression levels, respectively. The complete list of OS-related genes in this TCGA discovery cohort is provided in Supplementary Table S2. (e) Logarithm of the integrated HR for all 184 prognosis-related genes in the validation data sets. The complete list of these genes identified by meta-analysis is provided in Supplementary Table S3.