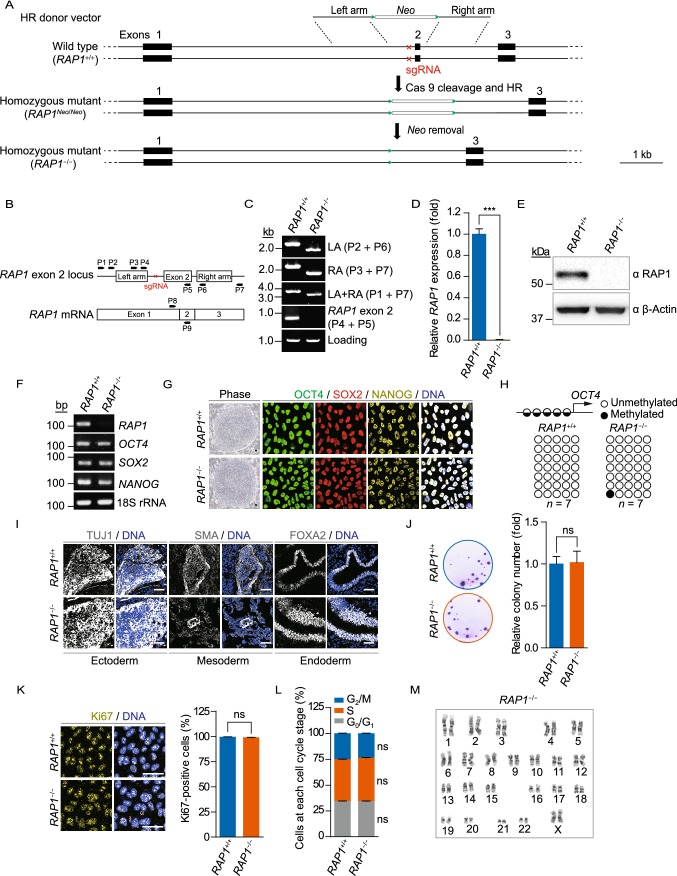
Figure 1.
Generation and characterization of RAP1−/− hESCs. (A) Schematic representation of the deletion of the exon 2 of RAP1 in hESCs via CRISPR/Cas9-facilitated HR. The green triangles represented FRT sites and the red cross demonstrated the region of sgRNA. (B) Schematic representation of the primers used for genomic PCR and qRT-PCR to confirm RAP1 knockout. (C) Genomic PCR analysis demonstrated that the exon 2 of RAP1 was deleted from the genome. LA and RA represented left and right homology arm, respectively. (D) qRT-PCR analysis demonstrated the deletion of RAP1 at the transcriptional level in RAP1−/− hESCs by primers P8 and P9. Data were presented as the mean ± SEM, n = 3. ***P < 0.001. (E) Western blotting analysis verified the absence of RAP1 in RAP1−/− hESCs. β-Actin was used as a loading control. (F) RT-PCR for pluripotency markers demonstrated comparable expression in WT and RAP1−/− hESCs. 18S rRNA was used as a loading control. (G) Representative brightfield and immunofluorescence micrographs of WT and RAP1−/− hESCs showed normal morphology of hESCs and comparable expression of pluripotency markers, respectively. Scale bar, 50 µm. (H) DNA methylation status of the OCT4 promoter in WT and RAP1−/− hESCs showed normal hypomethylation. (I) Immunostaining of representative markers of the three germ layers in teratomas formed by WT and RAP1−/− hESCs. Scale bar, 50 µm. (J) Clonal expansion analysis of WT and RAP1−/− hESCs. Data were presented as the mean ± SEM, n = 3. NS, not significant. (K) Immunostaining of the proliferation marker Ki67 in WT and RAP1−/− hESCs. Scale bar, 50 µm. Data were presented as the mean ± SEM, n = 6. NS, not significant. (L) Cell cycle analysis of WT and RAP1−/− hESCs. Data were presented as the mean ± SEM, n = 3. NS, not significant. (M) Karyotype analysis of RAP1−/− hESCs showed normal karyotype. n = 10