Abstract
Hantaviruses are zoonotic agents transmitted from small mammals, mainly rodents, to humans, where they provoke diseases such as Hemorrhagic fever with Renal Syndrome (HFRS) and its mild form, Nephropathia Epidemica (NE), or Hantavirus Cardio-Pulmonary Syndrome (HCPS). Hantaviruses are spread worldwide and monitoring animal reservoirs is of primary importance to control the zoonotic risk. Here, we describe the development of a pan-viral resequencing microarray (PathogenID v3.0) able to explore the genetic diversity of rodent-borne hantaviruses endemic in Europe. Among about 800 sequences tiled on the microarray, 52 correspond to a tight molecular sieve of hantavirus probes covering a large genetic landscape. RNAs from infected animal tissues or from laboratory strains have been reverse transcribed, amplified, then hybridized to the microarray. A classical BLASTN analysis applied to the sequence delivered through the microarray allows to identify the hantavirus species up to the exact geographical variant present in the tested samples. Geographical variants of the most common European hantaviruses from France, Germany, Slovenia and Finland, such as Puumala virus, Dobrava virus and Tula virus, were genetically discriminated. Furthermore, we precisely characterized geographical variants still unknown when the chip was conceived, such as Seoul virus isolates, recently emerged in France and the United Kingdom.
Subject terms: Biological techniques, Genotyping and haplotyping, High-throughput screening
Introduction
Hantaviruses are enveloped tri-segmented negative-stranded RNA viruses belonging to the Hantaviridae family, Bunyavirales order, according to the 2018 classification of the International Committee on Taxonomy of Viruses (ICTV)1. Hantaviruses are distributed worldwide, their animal reservoirs are rodents, insectivores or bats (orders Rodentia, Soricomorpha, Chiroptera). When rodent-borne orthohantaviruses are transmitted to humans through aerosolized excreta of infected rodent reservoirs, they can provoke Hemorrhagic Fever with Renal Syndrome (HFRS), its mild form Nephropathia Epidemica (NE), or Hantavirus Cardio-Pulmonary Syndrome (HCPS), mainly depending on the virus species and its genotype/strain. In Europe, cases of HFRS associated with Dobrava-Belgrade orthohantavirus (DOBV) and Seoul orthohantavirus (SEOV), and cases of NE associated with Puumala orthohantavirus (PUUV), are continuously reported in endemic areas such as the Balkans, the Fennoscandia region, Germany, Belgium, The Netherlands, France, and more recently, United Kingdom2–8. Tools to investigate the genetic diversity of hantaviruses in their animal reservoirs along their geographical distribution are particularly needed to better understand the epidemiology and predict hantavirus disease outbreaks in humans, and to set up appropriate public health measures9–14.
The recent development of molecular methods without an a priori hypothesis has brought a critical benefit in the field of diagnosis and research on infectious diseases. They can permit to obtain the sequence of a pathogen present in a biological sample, in absence of specific probes required for classic PCR or hybridization15–23. Among them, Next Generation Sequencing (NGS) have become widely used for identification and whole genome sequencing of novel animal or zoonotic pathogens and metagenome analysis22,23. These techniques are more accessible in terms of equipment and cost, but still require a complex downstream bioinformatics analysis which may represent a cumbersome step demanding a specific expertise. On the other hand, resequencing microarray do not demand a sophisticated analysis for interpretation. Indeed, the sequence obtained following hybridization of the amplified genetic material on the chip is used without any intermediate step for both BLASTN enquiry and phylogenetic analysis. Although the random amplification does not allow to specifically concentrate the viral material present in the specimen compared to classic or multiplex PCR assays, the following hybridization step on the resequencing microarray further improves the sensitivity and the specificity of the assay. In addition, the assay has the advantage of tolerating critical divergence levels, up to 20%, permitting a precise characterization from a short stretch of about 20 detected nt20, that can hardly be achieved when few reads are obtained following NGS assays. Resequencing microarray has been broadly used over the last ten years, for the detection and identification of emerging agents, such as monkeypox virus17,18, pandemic influenza viruses19, hemorrhagic fever viruses20, rhabdoviruses21. Several steps of the assay can be performed in the field (mobile laboratory) including sample preparation, random amplification by Phy29 (stable at room temperature) up to hybridization on the microarray. A specific equipment (fluidic station/scanner) is only required for the post-hybridization analysis and scanning.
PathogenID is a collaborative effort of teams of Institut Pasteur that combined expertise to develop 3 generations of resequencing microarrays for the detection of different emerging pathogens, bacteria and viruses, during public health urgencies and for research studies. The 1st and 2nd generations of the microarray contained only a limited number of viral sequences, 46 (PathogenID v1.0) and 126 (PathogenID v2.0), respectively. In particular, only sequences of six prototype orthohantaviruses, mainly associated with human diseases, were included: PUUV, DOBV, SEOV, Hantaan orthohantavirus (HTNV), Sin Nombre orthohantavirus (SNV), Andes orthohantavirus (ANDV)17,20. The targeted sequence corresponded to part of the large (L) segment coding for the RNA-dependent RNA-polymerase (RdRp), reputed to be the most conserved region of the genome24. While PathogenID v2.0 has been validated for detection of DOBV, SEOV, HTNV, ANDV and SNV in infected Vero E6 cells, it was not for PUUV, yet the most commonly circulating orthohantavirus in Europe20.
Therefore, the 3rd generation resequencing microarray (PathogenID v3.0), employed in the present cooperative study at the European scale has been specialized for virus detection and contains more than 800 viral sequences. This pan-viral resequencing microarray covers many viral pathogens critical for both animal and public health and in particular most of the known circulating species and variants of zoonotic viruses. Among those sequences, 52 were strategically chosen to cover the diversity of hantaviruses, in particular the most frequent rodent-borne species that are the only one reported today to have a zoonotic potential. The small (S) segment of the genome encoding the nucleocapsid (N) protein was chosen as target to design the probes since there are more efficient to discriminate variants within a species; in addition more reference sequences were present in GenBank at the moment of the pan-viral chip conception.
The present manuscript illustrates the use of the PathogenID v3.0 resequencing microarray to map the genetic diversity of several endemic hantaviruses mainly associated with human disease in Europe, such as PUUV, DOBV, including genotypes Dobrava and Saarema25, TULV, SEOV, Topografov hantavirus (TOPV) according to their geographical distribution.
Methods
Design of PathogenID v3.0 pan-viral resequencing microarray
The objective for the conception of the 3rd generation resequencing microarray (PathogenID v3.0) was to reach the widest coverage in virus diversity of both medical and veterinarian interest. Due to the technical limits of the microarray, we selected the minimum number of probes for each viral species/variant, in order to include the highest number of sequences. For each viral family, the included probes were chosen taking into account the reference sequences published in GenBank and our experience from the earlier generations of the microarray (PathogenID v1.0 and PathogenID v2.0)17–21.
PathogenID v3.0 contains 838 sequences including virus prototypes and their variants belonging to different families (complete list available upon request to the authors). Regarding hantaviruses, 52 N protein encoding S segment partial sequences were included (Table 1; Supplement 1), according to the known ICTV taxonomy in force when we designed the chip (e.g. choice of the probe sequences). The length of the tiled sequences was: i) 425 nucleotides (nt) for those available in GenBank, with the exception of seq234 (251 nt); ii) 303 nt for those sequenced in the laboratory during previous analyses and not published at the moment of the study (Table 1; Supplement 1).
Table 1.
List of 52 hantavirus sequences fixed on the panviral chip PathogenID v3.0.
| Family Hantaviridae (Species) | Isolate/Variant/strain Description* | GenBank Accession number | Sequence size (N segment size: nt-nt position)§ | Position chip# |
|---|---|---|---|---|
| Puumala orthohantavirus | CG14444 | AJ277075 | 425 (1837: 395–819) | 222 |
| CG 13891 | U22423 | 425 (1847: 395–819) | 223 | |
| Bavaria CG9/04 | AY954722 | 425 (1722: 392–816) | 224 | |
| Sotkamo NC 005224 | X61035 | 425 (1830: 395–819) | 225 | |
| Puu/Kazan_ | Z84204 | 425 (1826: 395–819) | 226 | |
| Umea/hu | AY526219 | 425 (1829: 395–819) | 227 | |
| PUU/Ernstbrunn/Cg641/1995 | AJ888752 | 425 (1852: 395–819) | 228 | |
| Berkel | L36943 | 425 (1068: 224–648) | 229 | |
| Couvin/59Cg/97 | AJ277034 | 425 (1839: 395–819) | 230 | |
| Pallasjarvi/63Cg/98 | AJ314597 | 425 (1827: 395–819) | 231 | |
| Munga/Mg16/05 | GQ339487 | 425 (1854: 395–819) | 232 | |
| PUU/Mignovillard/CgY02/2005 | AM695638 | 425 (1851: 395–819) | 233 | |
| France/Perpignan1999 | AY101391 | 251 (251: 1–251)° | 234 | |
| Fusong 900–06 | EF488806 | 425 (1302: 353–777) | 235 | |
| CH-214 Franche Comté | KT247596^ | 425 (1302: 353–777) | 236 | |
| AR-21 Charleville Meziéres | unpub^ | 425 (1775: 364–786) | 237 | |
| 167-2 Charleville Meziéres | unpub^ | 303 (1302: 352–654) | 238 | |
| 167-4 Charleville Meziéres | unpub^ | 303 (1302: 352–654) | 239 | |
| 178-2 Charleville Meziéres | unpub^ | 303 (1302: 352–654) | 240 | |
| 180-78 Charleville Meziéres | unpub^ | 303 (1775: 362–664) | 241 | |
| RU-11 Ruminy Troyes | KY364996^ | 303 (1785: 362–664) | 242 | |
| OR-52 Orleans | KT247595^ | 303 (1302: 352–654) | 243 | |
| Sin Nombre orthohantavirus | Convict Creek 107 | L33683 | 425 (2083: 395–819) | 244 |
| Dobrava-Belgrade orthohantavirus | Dobrava-Belgrade | L41916 | 425 (1670: 388–812) | 245 |
| Esl/81Aa/01 | AY533120 | 425 (1697: 388–812) | 246 | |
| Saaremaa orthohantavirus | Saaremaa/Lolland/Aa1403/2000 | AJ616854 | 425 (1673: 388–812) | 247 |
| Kenkeme orthohantavirus | MSB148794 | GQ306148 | 425 (1640: 403–827) | 248 |
| Hantaan orthohantavirus & unclassified hantaviruses | 1980471 | M14626 | 425 (1696: 389–813) | 249 |
| KY | GU140098 | 425 (1699: 389–813) | 250 | |
| Z10 | EF533944 | 425 (1701: 389–813) | 251 | |
| AA1719 | AF427319 | 425 (1643: 366–790) | 252 | |
| CGHu1 | EU092218 | 425 (1681: 389–813) | 253 | |
| Q32 | AB027097 | 425 (1635: 353–777) | 254 | |
| Seoul orthohantavirus | Z37 | AF187082 | 425 (1754: 395–819) | 255 |
| Hantaan orthohantavirus | Nc167 | AB027523 | 425 (1654: 354–778) | 256 |
| Seoul orthohantavirus | L99 | AF488708 | 425 (1290: 353–777) | 257 |
| Seoul orthohantavirus | 80-39 | NC005236 | 425 (1769: 395–819) | 258 |
| Asama orthohantavirus | N10 | EU929072 | 425 (1710: 371–795) | 259 |
| Nova mobatvirus | MSB95703 | FJ539168 | 425 (1839: 405–829) | 260 |
| Prospect Hill orthohantavirus | PHV 1980485 | M34011 | 425 (1675: 395–819) | 261 |
| Soochong virus (unclass.) | SC-1 | AY675349 | 425 (1695: 388–812) | 262 |
| Tula orthohantavirus | Sennickerode Sen05/204 | EU439950 | 425 (1700: 395–819) | 263 |
| Tula/Kosice144/Ma/95 | Y13979 | 425 (1833: 395–819) | 264 | |
| Tula/Moravia/5293 Ma/94 | Z48574 | 425 (1832: 395–819) | 265 | |
| MG23/Omsk | AF442621 | 425 (1758: 353–777) | 266 | |
| Thottapalayam thottivirus | Thottapalayam 1980493 | AY526097 | 425 (1530: 450–874) | 267 |
| Andes orthohantavirus | Chile-9717869 | AF291702 | 425 (1871: 395–819) | 268 |
| NK104619 | EU241691 | 425 (927: 189–613) | 269 | |
| Sin Nombre orthohantavirus | NM R11 | L37904 | 425 (2060: 395–819) | 270 |
| New York orthohantavirus | RI-1 44755 | U09488 | 425 (2078: 395–819) | 271 |
| Choclo virus orthohantavirus | 169173 | DQ285046 | 425 (1972: 395–819) | 272 |
| Laguna Negra orthohantavirus | 510B 1980476 | AF005727 | 425 (1904: 395–819) | 273 |
All sequences are partially targeting the S segment encoding the hantavirus nucleocapsid (N) protein. Complementary information is found in Supplement 1.
*Description of the isolate/strain as indicated in GenBank or according to laboratory nomenclature for experimentally obtained sequences.
§Position according to the corresponding sequence available in GenBank.
°Sequence corresponding to a different region of the S gene.
#The number is referred to the position on the pan-viral chip PathogenID v3.0 (total number: 838 sequences).
^unpublished at the moment of the study.
Upon sequence selection, PathogenID v3.0 was manufactured by Affymetrix (Santa Clara, California) according to their high density resequencing approach, based on the use of stepwise overlapping 25 nt long probes, the first covering position 1–25 of the tiled sequence, the second position 2–26, etc.: each probe comprises a set of 4 different alleles differing in the central 13th position (e.g. A, C, G or T)16,17. A total of 2.5 millions of 25 mer oligonucleotides were tiled on the microarray PathogenID v3.0.
Hantavirus plasmids
Synthetic plasmids containing the S segment of PUUV, the most endemic hantavirus in Europe (strain Sotkamo 2009), HTNV (strain 76/118), TULV (strain Moravia) were used in a preliminary assay to evaluate the potential of the microarray for hantavirus detection, prior testing the field animal samples. PUUV plasmid was taken as reference and tested alone and in pool with the HNTV and TULV plasmids.
Laboratory strains
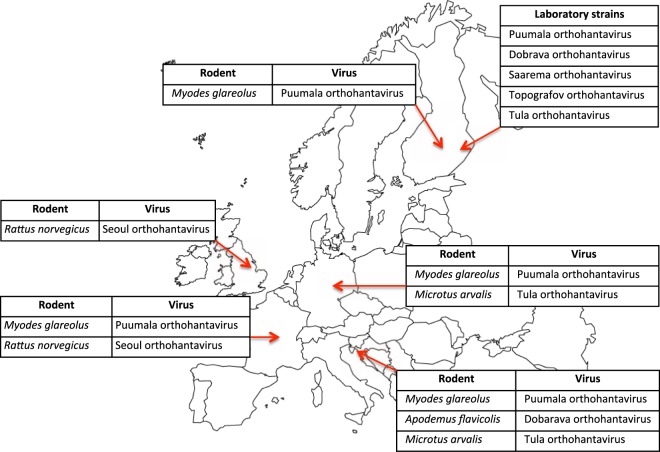
Laboratory prototype strains of PUUV, DOBV (genotypes Dobrava and Saaremaa) TULV and TOPV isolated on Vero E6 cells in BSL3 containment were provided by the virology participant laboratory in Finland26–30 (Fig. 1).
Figure 1.
Cooperative work for genetic characterization study of endemic hantaviruses in Europe. Hantaviruses from rodents: Bank vole (Myodes glareolus), Common vole (Microtus arvalis), Yellow-necked mouse (Apodemus flavicolis), Striped field mouse (Apodemus agrarius), and Norway rat (Rattus norvegicus) captured in different endemic areas in Europe were genetically characterized using a resequencing pan-viral chip during a cooperative work among European research institutes: France, Germany, Scandinavia, Balkans, United Kingdom. Laboratory viral strains were also obtained from one of the participant laboratory. The tested RNA included: (i) RNA extracted from tissues (e.g. lung) originating from infected wild rodents; (ii) RNA extracted from supernatants from inoculated cells (Vero E6).
Endemic hantaviruses from wild rodents captured in Europe
Hantavirus rodent reservoirs of the Bank vole (Myodes glareolus), Common vole (Microtus arvalis), Yellow-necked mouse (Apodemus flavicolis), Striped field mouse (Apodemus agrarius), and Norway rat (Rattus norvegicus) were previously captured in different hantavirus endemic areas in Europe: France, Germany, Finland, Slovenia, and United Kingdom30–41. Figure 1 illustrates the capture areas and rodent species as well as the Institutes participating to the study and providing positive animals.
Lung samples were kept at −80 °C in RNAlater (Ambion, Thermo Fisher Scientific, Waltham, USA) prior to RNA extraction (QiaAmp Viral RNA Kit, Qiagen, Hilden, Germany). Hantavirus positive RNAs, routinely confirmed by the reference laboratories by RT-PCR30–41, were employed for exploring their genetic diversity by resequencing microarray.
France
PUUV, the most frequent hantavirus circulating in France, was obtained from bank voles samples captured in France in 2011 in the Ardennes region31, SEOV (Lyon strain) from Norway rats39.
Finland for Fennoscandia region
Lung tissues originating from PUUV-positive bank voles captured in Konnevesi in 2008 were used30.
Germany
PUUV and TULV RNAs originating from lungs of animal reservoirs captured across Germany32–36 were used.
Balkan region
PUUV, TULV, DOBV RNAs originating from lungs were obtained from rodents captured in Slovenia37,38.
United Kingdom
SEOV (Cherwell strain) obtained from organs of domestic rats in United Kingdom (UK) was used40,41.
All experiments were performed in accordance with relevant guidelines and regulations. RNAs extracted from tissue sample from wild animals were received from various Hantavirus Reference Centers in Europe (Fig. 1) through the EU program EVA (European Virus Archive - n° 228292) which facilitates access to virus/tissue library under MTA (Material Transfer Agreement). All of them have been previously published in peer-reviewed literature. All handling procedures of captured rodents followed the regulations of each respective country. The species studied are not protected and all efforts were made to minimize animal suffering.
Genetic detection and phylogeographical characterization of hantaviruses by the pan-viral chip PathogenID v3.0
The experimental procedure for hantavirus typing includes the following steps, detailed in previous works19,20 (Supplement 2).
RNA extraction
RNA was extracted using the QIAamp Viral RNA Extraction kit (Qiagen) from both animal organ homogenates or cell supernatants. cDNA was synthetized by Superscript III system (Invitrogen, Thermo Fisher).
Random amplification
Genetic material, either plasmid DNA, or cDNA from animal organs or cell supernatants, was amplified by WGA (Whole Genome Amplification) and WTA (Whole Transcriptome Amplification) approaches, respectively, using ϕ29 polymerase-mediated random amplification (Qiagen), followed by a ligation step17.
Hybridization on the microarray and sequence detection
Amplified products were hybridized overnight at 45 °C on the microarray PathogenID v3.0 after fragmentation and labeling using GeneChip Resequencing Reagent Kit (Affymetrix). Chips were then subjected to washing, fluorescence detection and scanning using the Affymetrix equipment (Wash Control, Scan Control). Resequencing analysis was performed using the software GSEQ. 4.1 (Affymetrix). For each of the 52 hantavirus sequences fixed on the chip (positions seq222 to seq273), an output sequence was obtained in a .txt format, with determined (A, G, T or C) or non-determined (N) positions (example in Supplement 2). Significant sequences obtained were used for Call Rate calculation and BLASTN analysis.
Call Rate calculation
Call Rate (CR) was calculated as the ratio (%) between the number of determined (‘called’) nucleotides (e.g. A, G, T, C) following hybridization and the total number of nucleotides for each tiled sequence (e.g. 425, 251 or 303 nt, for hantaviruses).
BLASTN analysis
For samples presenting a minimum stretch of informative sequence20, the entire raw sequence was submitted to BLASTN (Basic Local Alignment Search Tool Nucleotide) query [National Center for Biotechnology Information, National Institute of Health], to interrogate all the sequences present in GenBank. In case of positive BLASTN result, scores, identity (%), query coverage (%) and e-value were taken into account.
Phylogenetic analysis
Phylogenetic analysis was performed for PUUV (as representative of European orthohantavirus) by using (i) the reference sequences (S segment) available on GenBank, (ii) the sequences tiled on the chip, (iii) the sequences corresponding to the tested hantaviruses, when known.
Firstly, a phylogenetic tree was constructed with the complete coding part of reference sequences by the maximum-likehood method (ML) with PhyML v3.0, implemented in Seaview (v.4.6.1) under the most appropriate nucleotide substitution model as determined by SMS program (available online at http://www.atgc-montpellier.fr/sms/)42. Branch supports were evaluated by approximate likelihood-ratio test (aLRT SH-Like). Then, short sequences of the chips and of tested hantaviruses were placed in this backbone tree using RAxML available online on the CIPRES portal at (http://www.phylo.org). Branch supports of these phylogenetic placements were evaluated by the rapid bootstrap procedure with MRE-based Bootstopping criterion as highly recommended on the online software version.
For each sample giving a positive result following hybridization, BLASTN analysis of the resulting sequence, was pointed out on the phylogenetic tree, and compared to the sequences having permitted the detection and genetic characterization.
Results
Initial validation of the 3rd generation pan-viral resequencing microarray for hantavirus detection and genetic characterization
The performance in hantavirus detection and genetic characterization of the resequencing chip Pathogen ID v3.0 was first evaluated using plasmids encoding prototype hantavirus sequences (PUUV, HTNV, TULV). The PUUV plasmid encompassing the N protein coding region (1831 nt) of the reference Sotkamo strain 2009 was hybridized to Pathogen ID v3.0 which includes 22 PUUV S segment sequences (seq222 to seq243) (Table 1; Supplement 1). For each of these sequences, Table 2 summarizes (i) the calculated sequence similarity (%) with the tested PUUV Sotkamo strain sequence; (ii) the percentage of correctly identified nucleotides (CR) following its hybridization to the chip and (iii) the result of the BLASTN analysis performed with the obtained raw sequences (Supplement 3).
Table 2.
Validation of the Pathogen ID v3.0 resequencing microarray using a nucleocapsid (N) protein encoding plasmid of the PUUV prototype strain Sotkamo.
| PUUV sequence on the chip (position) | Sequence similarity to PUUV Sotkamo (nbr identical nt/total nbr nt) - (%) | BLASTN identification | Call Rate (CR) (%) |
|---|---|---|---|
| seq222 | 344/422 - 81.5 | yes: Sotkamo | 23.9 |
| seq223 | 335/422 - 79.4 | no | 23.7 |
| seq224 | 346/425 - 81.4 | no | 18.9 |
| seq225* | 425/425* - 100* | yes: Sotkamo | 97.7 |
| seq226 | 351/425 - 82.6 | no | 26.2 |
| seq227 | 357/424 - 84.2 | yes: Sotkamo | 39.6 |
| seq228 | 346/423 - 81.8 | yes: Sotkamo + FIN variants | 22.7 |
| seq229 | 346/425 - 81.4 | no | 19.4 |
| seq230 | 347/422 - 82.2 | yes: Sotkamo | 33.9 |
| seq231 | 387/423 - 91.5 | yes: Sotkamo | 75.0 |
| seq232 | 353/424 - 83.2 | yes: Sotkamo + FIN variants | 34.4 |
| seq233 | 342/424 - 80.7 | yes: Sotkamo + FIN variants + others° | 19.7 |
| seq234# | # | yes: Sotkamo | 62.1 |
| seq235 | 323/422 - 76.5 | no | 7.0 |
| seq236 | 343/424 - 80.9 | yes: Sotkamo + FIN variants + others° | 19.9 |
| seq237 | 348/423 - 82.3 | yes: Sotkamo | 35.2 |
| seq238 | 244/299 - 81.6 | no | 24.7 |
| seq239 | 247/299 - 82.6 | yes: Sotkamo | 34.4 |
| seq240 | 249/299 - 83.3 | yes: Sotkamo | 38.3 |
| seq241 | 242/299 - 80.9 | no | 26.9 |
| seq242 | 245/299 - 81.9 | yes: Sotkamo + FIN variants + others° | 30.8 |
| seq243 | 246/301 - 81.7 | no | 25.1 |
A plasmid containing the N protein encoding sequence of the PUUV Sotkamo strain was used to evaluate the range of detection of the Pathogen ID v3.0 resequencing microarray. Sequence identity (%) was calculated by aligning the tested Sotkamo PUUV sequence against each PUUV sequence tiled on the array (seq222–seq243). Following hybridization of the Sotkamo plasmid on the chip, each significant output sequence was evaluated by its Call Rate (CR) value (% of the correctly determined nucleotides of the sequence following hybridization) and the result(s) of its analysis by BLASTN. Complementary information is given in Supplement 3 and Fig. 2.
*Sotkamo sequence tiled on the chip.
#different region of the S segment tiled on the chip.
°others: PUUV variants other than Sotkamo and Finnish (FIN) variants.
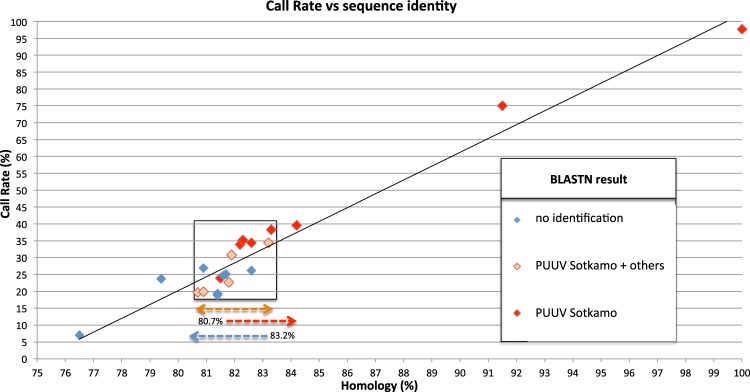
It clearly appeared that the CR values were proportional to the sequence similarity between the tested sequence and the tiled ones (Table 2). From 100% (seq225, Sotkamo itself) to 91.5% (seq231) of identity, the CR was very high (97.7% to 75%, resp.) and the BLASTN identified the Sotkamo strain without ambiguity. Down to 82.2% of sequence identity (seq230), the CR remained above 34%, still designing Sotkamo in priority by BLASTN, although some tiled sequences already hesitated in precise identification (seq232) or even failed in identification (seq226). Down to 82% of homology, the CR decreased dramatically and the tested sample failed to be identified in some cases. Plotting of the sequence identity to the CR confirmed these observations (Fig. 2): below 80.7% of identity between the tested and the tiled sequences the microarray becomes inefficient for specific detection; the window between 80.7% and 83.2% of identity is critical and versatile between no detection, detection with unspecific determination, and precise genetic characterization of the tested sequence; higher than 83.2% of identity, the microarray identifies precisely the tiled sequence.
Figure 2.
Performance of the Pathogen ID v3.0 resequencing microarray to detect and identify hantavirus sequences. A plasmid encoding the nucleocapsid (N) protein of the PUUV prototype strain Sotkamo was used to evaluate the range of detection of the Pathogen ID v3.0 resequencing microarray. This figure compares the calculated sequence identity (%) between the tested sequence (PUUV Sotkamo) and each PUUV sequences tiled on the microarray with the respective Call Rate (CR, i.e. % of determined nucleotides) obtained after hybridization. Colours, values and arrows outline a window of sequence identity (%) for BLASTN results obtained from each output sequence following hybridization: from no detection/identification (blue), to general PUUV characterization (Sotkamo + others, orange) to precise and exclusive characterization (Sotkamo, red). Complementary information is described in Table 2 and Supplements 3–4.
We verified that simultaneous detection was possible when plasmids containing the N protein coding region of the three hantavirus reference strains (PUUV Sotkamo 2009, TULV Moravia, HTNV 76/118) were mixed in pool (Supplement 3–4). The CR values of PUUV output sequences (seq222–seq243) were even higher (e.g. more determined nucleotides) for the pool of the three viruses, most likely due to cross-contribution from the three viruses in hybridizing the same sequence tiled on the chip, thus producing better results in BLASTN analysis (Supplements 3–4).
Mapping genetic diversity of hantaviruses circulating in Europe
The same technology validated with plasmids encoding hantavirus N protein was applied to tissue samples of PUUV, TULV, DOBV, SAAV, SEOV, TOPV infected rodents originating from different endemic areas in Europe as well as to supernatants of cells infected with laboratory strains (Tables 3–7; Supplement 5). Using RNA extracted from lung and liver of PUUV RNA positive bank voles captured in the Ardennes region of France in 201131 we first observed that, at comparable viral RNA titer (Ct value), lung was more performing for hantavirus investigation on the chip (data not shown). Therefore, lung derived RNAs were priorized for further investigation.
Table 3.
Detection and genetic characterization of Puumala orthohantaviruses (PUUV) from various endemic area in Europe: PUUV isolates or laboratory strain were hybridized to the hantavirus sequences tiled on the PathogenID v3.0 resequencing microarray (seq222 to seq273; Table 1; Supplement 1). For each tested sample, the tiled sequences giving a significant signal are listed and the Call Rate (CR, i.e. % of correctly determined nucleotides) indicated. The corresponding raw sequences were subjected to BLASTN enquiry. The number (nbr) of BLASTN identical results is indicated; the most related sequence is indicated in italics and a maximum of the first 3 closer sequences are listed. “Total Score” evaluates the overall quality of the alignment by BLASTN. Complementary data including all sequences obtained following BLASTN analysis and further details are described in Supplement 5.
| Tested virus (geographical origin) | Tiled sequence allowing detection (position on the chip) | BLASTN identification | Score (total) | Call rate (%) | |
|---|---|---|---|---|---|
| nbr | sequences | ||||
| PUUV 87 (France) | 222: PUUV CG14444 | 3 | Ardennes_2011_87; Ardennes/Mg75/2011; Ardennes/Mg156/2011 | 226 | 65.6 |
| 223: PUUV CG13891 | 1 | Ardennes_2011_87 | 89.1 | 35.9 | |
| 230: Couvin/59Cg/97 | 3 | Ardennes_2011_87; Ardennes/Mg75/2011; Ardennes/Mg156/2011 | 351 | 73.8 | |
| 237: AR-21 Charlesville_Mezieres | 3 | Ardennes_2011_87; Ardennes/Mg75/2011; Ardennes/Mg156/2011 | 268 | 67.8 | |
| 238: 167-2 Charlesville-Mezieres | 6 | Ardennes_2011_87; Ardennes/Mg75/2011; Ardennes/Mg156/2011 | 108 | 52.7 | |
| 239: 167-4 Charlesville-Mezieres | 3 | Ardennes_2011_87; Ardennes/Mg75/2011; Ardennes/Mg156/2011 | 288 | 78.5 | |
| 240: 178-2 Charlesville-Mezieres | 3 | Ardennes_2011_87; Ardennes/Mg75/2011; Ardennes/Mg156/2011 | 268 | 77.4 | |
| 241: 180-78 Charlesville-Mezieres | 3 | Ardennes_2011_87; Ardennes/Mg75/2011; Ardennes/Mg156/2011 | 239 | 73.8 | |
| PUUV 153 (France) | 222: PUUV CG14444 | 3 | Ardennes_2011_87; Ardennes/Mg75/2011; Ardennes/Mg156/2011 | 488 | 83.0 |
| 223: PUUV CG13891 | 3 | Ardennes_2011_87; Ardennes/Mg75/2011; Ardennes/Mg156/2011 | 141 | 52.3 | |
| 229: Berkel_L36943 | 28 | Ardennes_2011_87; Ardennes/Mg75/2011; Ardennes/Mg156/2011 | 44.6 | 26.4 | |
| 230: Couvin/59Cg/97 | 3 | Ardennes_2011_87; Ardennes/Mg75/2011; Ardennes/Mg156/2011 | 581 | 89.5 | |
| 234: Perpignan 1999 | 1 | Ardennes/Mg156/2011 | 150 | 64.3 | |
| 237: AR-21 Charlesville_Mezieres | 3 | Ardennes_2011_87; Ardennes/Mg75/2011; Ardennes/Mg156/2011 | 547 | 87.5 | |
| 238: 167-2 Charlesville-Mezieres | 3 | Ardennes_2011_87; Ardennes/Mg75/2011; Ardennes/Mg156/2011 | 156 | 68.1 | |
| 239: 167-4 Charlesville-Mezieres | 3 | Ardennes_2011_87; Ardennes/Mg75/2011; Ardennes/Mg156/2011 | 392 | 88.9 | |
| 240: 178-2 Charlesville-Mezieres | 3 | Ardennes_2011_87; Ardennes/Mg75/2011; Ardennes/Mg156/2011 | 408 | 91.0 | |
| 241: 180-78 Charlesville-Mezieres | 3 | Ardennes_2011_87; Ardennes/Mg75/2011; Ardennes/Mg156/2011 | 345 | 84.2 | |
| PUUV Mu557 Gilserberg/10 (Germany) | 223: PUUV CG13891 | 27 | Mu557Gilserberg/10 | 44.6 | 33.4 |
| 230: Couvin/59Cg/97 | 1 | Mu557Gilserberg/10 | 121 | 34.2 | |
| 233: Mignovillard/CgY02/2005 | 1 | Mu557Gilserberg/10 | 153 | 35.4 | |
| 234: Perpignan1999 | 2 | PUUV/NL/Mg591/2008; Heidelberg/hu | 80.6 | 54.2 | |
| 236: CH-214_Franche_Comté | 1 | Mu557Gilserberg/10 | 57.2 | 28.7 | |
| 237: AR-21_Charlesville_Mezieres | 1 | Mu557Gilserberg/10 | 73.4 | 32.4 | |
| 238: 167-2Charleville_Mezieres | 1 | Mu557Gilserberg/10 | 57.2 | 41.2 | |
| 239: 167-4Charleville_Mezieres | 1 | Mu557Gilserberg/10 | 57.2 | 38.0 | |
| 240: 178-2Charleville_Mezieres | 7 | Mu557Gilserberg/10 | 53.6 | 40.9 | |
| 241: 180-78Charleville_Mezieres | 1 | Mu557Gilserberg/10 | 90.9 | 41.6 | |
| 242: RU-11Ruminy-Troies | 2 | Mu557Gilserberg/10; H290 Fulda/10 | 64.4 | 30.8 | |
| PUUV Mu2232 Bramsche/09 (Germany) | 224: Bavaria CG9/04 | 34 | Mu2232Bramsche/09 | 60.8 | 31.2 |
| 229: Berkel_L36943 | 6 | KS10/3078; KS14/873; KS14/715 | 158 | 39.7 | |
| 233: Mignovillard/CgY02/2005 | 2 | KS10/3078; KS14/778 | 94.5 | 28.4 | |
| 234: Perpignan 1999 | 2 | Mu/07/1219; Mu362Osnabrueck/05 | 48.2 | 41.0 | |
| 242: RU-11RuminyTroyes | 11 | Mu2232Bramsche/09 | 60.8 | 30.5 | |
| 243: OR-52_Orleans | 42 | Mu2232Bramsche/09 | 44.6 | 30.1 | |
| PUUV Mu978 Weissach/10 (Germany) | 222: CG14444 | 6 | Mu978Weissach/10 | 129 | 42.4 |
| 224: Bavaria CG9/04 | 3 | Mu978Weissach/10 | 172 | 48.6 | |
| 234: Perpignan1999 | 1 | Heidelberg/hu | 132 | 59.5 | |
| 240: 178-2Charleville-Mezieres | 6 | Mu978Weissach/10 | 89.7 | 45.5 | |
| 241: 180-78Charleville-Mezieres | 6 | Mu978Weissach/10 | 87.8 | 40.1 | |
| 242: RU-Ruminy-Troyes | 6 | Mu978Weissach/10 | 75.2 | 39.4 | |
| PUUV laboratory strain Sotkamo | 222: CG14444 | 2 | Sotkamo 2009; Puumala virus genomic RNA for nucleocapsid protein | 51.8 | 23.9 |
| 225: Sotkamo_NC_005224 | 2 | Sotkamo 2009; Puumala virus genomic RNA for nucleocapsid protein | 673 | 96.5 | |
| 227: Umea | 2 | Sotkamo 2009; Puumala virus genomic RNA for nucleocapsid protein | 51.8 | 28.9 | |
| 228: Ernstbrunn/Cg641/1995 | 6 | Sotkamo 2009 | 69.8 | 22.7 | |
| 229: Berkel_L36943 | 10 | Sotkamo 2009 | 44.6 | 21.2 | |
| 230: Couvin/59Cg/97 | 2 | Sotkamo 2009; Puumala virus genomic RNA for nucleocapsid protein | 50.0 | 30.4 | |
| 231: Pallasjarvi/63Cg/98 | 1 | Puumala virus N and Ns genes, segment S, genomic RNA, Sotkamo2009 | 361 | 70.1 | |
| 232: Munga/Mg16/05 | 5 | Puumala virus N and Ns genes, segment S, genomic RNA, Sotkamo2009 | 68.0 | 27.4 | |
| 233: Mignovillard/CgY02/2005 | 3 | Puumala virus N and Ns genes, segment S, genomic RNA, Sotkamo2009 | 51.8 | 15.5 | |
| 234: Perpignan1999 | 2 | Sotkamo2009; Puumala virus genomic RNA for nucleocapsid protein | 64.4 | 50.2 | |
| 236: CH-14_Franche_Comté? Jura | 3 | Sotkamo2009 | 55.4 | 18.9 | |
| 237: AR-21_Charleville_Mezières | 2 | Sotkamo 2009; Puumala virus genomic RNA for nucleocapsid protein | 46.4 | 31.2 | |
| 239: 167-4Charleville_Mezières | 2 | Sotkamo 2009; Puumala virus genomic RNA for nucleocapsid protein | 51.8 | 31.2 | |
| 240: 178-2Charleville_Mezières | 2 | Sotkamo 2009; Puumala virus genomic RNA for nucleocapsid | 44.6 | 30.1 | |
| 242: RU-11Ruminy-Troyes | 3 | Sotkamo 2009 | 60.8 | 23.3 | |
| PUUV 335 Konnevesi (variant A) (Finland) | 225: Sotkamo_NC_005224 | 1 | Puumala virus strain PUUV/Konnevesi/Mg_O78A/2005 segment S | 112 | 44.4 |
| 231: Pallasjarvi/63Cg/98 | 2 | PUUV/Konnevesi/Mg_O78A/2005 | 127 | 38.6 | |
| 234: Perpignan1999 | 48 | PUUV/Konnevesi/Mg_O78A/2005 segment S | 46.4 | 25.5 | |
| 242: RU-11Ruminy-Troyes | 5 | Puumala virus strain PUUV/Konnevesi/Mg_O78A/2005 segment S | 55.4 | 13.6 | |
| PUUV Slovenia 8098 (Slovenia) | 225: Sotkamo_NC_005224 | 37 | Puumala virus isolate HtSi_339_p2012 nucleocapsid protein gene, partial | 53.6 | 16.2 |
| 226: Kazan_Z84204 | 19 | Puumala virus isolate HtSi_293_p2010 nucleocapsid protein gene, partial | 44.6 | 12.7 | |
| 228: Ernstbrunn/Cg641/1995 | 1 | Puumala virus isolate HtSi_293_p2010 nucleocapsid protein gene, partial | 167 | 55.9 | |
| 231: Pallasjarvi/63Cg/98 | 36 | Puumala virus isolate HtSi_339_p2012 nucleocapsid protein gene, partial | 73.4 | 28.2 | |
| 233: Mignovillard/CgY02/2005 | 41 | Puumala virus isolate HtSi_293_p2010 nucleocapsid protein gene, partial | 48.2 | 20.4 | |
| 234: Perpignan1999 | 5 | Puumala virus mRNA for nucleocapsid protein (N gene), strain Balkan-2 | 109 | 58.6 | |
| 242: RU-11Ruminy-Troyes | 7 | Puumala virus isolate HtSi_293_p2010 nucleocapsid protein gene, partial | 50.0 | 24.0 | |
Table 7.
Detection and genetic characterization of Topografov orthohantavirus (TOPV): the laboratory strain Topografov was hybridized to the hantavirus sequences tiled on the PathogenID v3.0 resequencing microarray (seq222 to seq273; Table 1; Supplement 1).
| Tested virus (geographical origin) | Tiled sequence allowing detection (position on the chip) | BLASTN identification | Score (total) | Call rate (%) | |
|---|---|---|---|---|---|
| nbr° | sequences | ||||
| Topografov laboratory strain | 228: Ernstbrunn/Cg641/1995 | 1 | Topografov hantavirus S segment gene for N protein | 44.6 | 15.0 |
| 234: Perpigna1999 | 12 | Topografov hantavirus S segment gene for N protein | 60.8 | 30.8 | |
| 235: Fusong90006 | 1 | Topografov hantavirus S segment gene for N protein | 50.0 | 29.4 | |
See legend of Table 3 for details on presented results.
Genetic characterization of Puumala viruses circulating in Europe
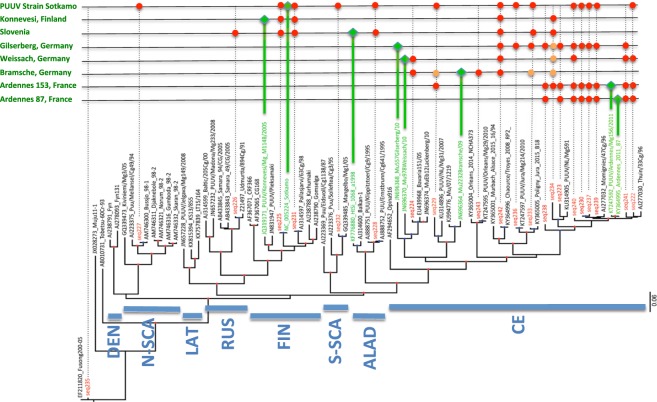
RNA extracts from supernatant of Vero E6 cells infected with the PUUV Sotkamo strain, and RNA extracts from lungs of seven PUUV-infected bank voles originating from France, Germany, Finland or Slovenia were individually hybridized to the 22 PUUV sequences (seq222 to seq243) tiled on the PathogenID v3.0 resequencing microarray. It is of note that only two of the tested samples had their exact sequence tiled on the chip: the PUUV Sotkamo strain (seq225) and the French Ardennes PUUV strain 87 (seq237). When a significant signal was detected, the corresponding raw sequence was subjected to BLASTN enquiry for genetic typing (Table 3). Figure 3 pictures the results in the context of a phylogenetic tree illustrating the currently known diversity of PUUV by combining references sequences available in GenBank, sequences tiled on the chip (in red) and sequences of the tested viruses (in green). An unequivocal determination of the correct geographical variant was observed for all the tested samples at least with one tiled sequence (red dots in Fig. 3), even when the corresponding sequence was not tiled itself on the chip. In very few cases (<8%, orange dots in Fig. 3) tiled sequences designated only an approximate origin, however always in the same genetic cluster.
Figure 3.
Phylogenetic analysis of European Puumala orthohantavirus (PUUV) genetically characterised by PathogenID v3.0 resequencing microarray. A phylogenetic tree backbone was first constructed from complete S segment coding sequences of reference hantaviruses available in GenBank using the maximum-likelihood method (ML) with PhyML v3.0, implemented in Seaview (v.4.6.1) with the most appropriate substitution model as determined by SMS program. Shorter sequences (tiled on the chip & tested viruses) were then placed in the backbone tree using RaxML. Nodes with branch support values > 0.8 are indicated by a red point (reference tree, aLRT branch support test) or by a blue point (phylogenetic placement of short sequence in the reference tree, rapid bootstrap procedure with MRE-based Bootstopping criterion). Scale bar represents the average number of substitutions per site. Under the tree are indicated the main geographical clusters of PUUV in Europe: CE (Central Europe); ALAD (Alpes-Adrian); S-SCA (South Scandinavia); FIN (Finland); RUS (Russia); LAT (Latvia); N-SCA (North Scandinavia); DEN (Denmark). The 22 PUUV sequences fixed on the chip (seq222 to seq243) are outlined in red, those of the 8 tested viruses in green. In the upper grid, the 8 tested Puumala viruses (the laboratory PUUV Sotkamo strain and 7 European geographical variants) are presented with: their exact position in the tree (green diamond); the tiled sequences having permitted their detection (significant CR) with exact (red circles) or close (orange circles) genetic identification by BLASTN. Complementary information is described in Table 3 and Supplement 5.
The French Ardennes PUUV lung-derived variants 87 and 153 were precisely identified by BLASTN not only using the very homologous French Ardennes PUUV tiled sequences (seq237-seq241), but also with tiled sequences from Belgium (seq222-seq223, seq230) or even from North-West Germany (seq229 for variant 153) more distant phylogenetically within the Central European (CE) clade. With the same logic, the three PUUV variants from Germany were precisely characterized by 91% (10/11; Gilserberg), 84% (5/6; Weissach) and 50% (3/6; Bramsche) of the hybridizing homologous and heterologous tiled sequences from the Central European clade (Table 3). More interestingly, the Konnevesi variant from Finland was exactly identified not only with tiled sequences from its specific clade (Finland, FIN) but also from the Central European (CE) clade. Equally, the variant 8098 from Slovenia (clade Alpes-Adrian, ALAD) was exactly identified with sequences from the ALAD clade, and also from the CE, FIN and Russia (RUS) clades. Finally, the laboratory PUUV Sotkamo strain was systematically identified by tiled sequences from almost all clades of the PUUV phylogenetic tree.
It is of note that these very precise characterisations could be obtained despite very low CR values, for example down to only 13% of defined nucleotides between the tested PUUV variant Slovenia 8098 and the tiled seq226 (Kazan_Z84204) (Table 3). It is explained by the exact determination of short fragments (stretches) of significant sequence (e.g. minimum of 15 nucleotides) that allow precise BLASTN identification despite poor CR values (Table 3; Supplement 5).
From Puumala virus to other orthohantaviruses circulating in Europe
The potential in detection and genetic characterization of the resequencing chip Pathogen ID v3.0 was further evaluated for variants and laboratory strains of other hantavirus species circulating in Europe, namely DOBV, TULV, SEOV and TOPV. For this purpose, 30 additional hantavirus sequences (seq244 to seq273) were tiled on the microarray.
For DOBV, RNA extracts from two cell supernatants infected with reference laboratory strains Belgrade and Saaremaa and RNA extracts from two Yellow-necked mice from Slovenia were tested (Table 4). All of them were exactly characterized: (1) by the DOBV (seq245-seq246) and/or SAAV (seq247) sequences tiled on the chip, as expected from the genetic similarity of both viruses; (2) more interestingly, by tiled sequences from more distant orthohantavirus species such as HTNV and Soochong virus. Significant results were also observed for TULV RNA extracts from one laboratory strain and three Common vole samples from Germany, Finland and Slovenia (Table 5). Following BLASTN, the correct sequence was either exclusively or at least dominantly characterized. Here again, precise detection and identification was possible with tiled sequences from very distant clades of vole-associated New world orthohantaviruses such as Prospect Hill or Sin Nombre orthohantaviruses (Table 5). Finally the SEOV present in RNA extracts from Norway rats from France and UK, although unknown when the chip was designed, was perfectly identified by heterologous SEOV tiled sequences (Table 6). Similarly for RNA extracts from a TOPV laboratory strain, in absence of the corresponding sequence tiled on the chip, the exact characterization was achieved with PUUV tiled sequences (seq228, seq234, seq325) even at low CR of 15% (Table 7).
Table 4.
Detection and genetic characterization of Dobrava orthohantaviruses (DOBV) from various endemic area in Europe: DOBV isolates or laboratory strains were hybridized to the hantavirus sequences tiled on the PathogenID v3.0 resequencing microarray (seq222 to seq273; Table 1; Supplement 1).
| Tested virus (geographical origin) | Tiled sequence allowing detection (position on the chip) | BLASTN identification | Score (total) | Call rate (%) | |
|---|---|---|---|---|---|
| nbr° | sequences | ||||
| DOBV-laboratory strain | 245: Belgrade L41916 | 1 | DOBV/Ano-Poroia/Afl9/1999 | 293 | 70.8 |
| Saaremaa laboratory strain | 245: Belgrade L41916 | 1 | DOB/Saaremaa/160 V | 137 | 44.1 |
| 247: Saaremaa virus AJ616854 | 1 | DOB/Saaremaa/160 V | 145 | 48.4 | |
| 256: Hantavirus NC167 AB027523 | 9 | DOB/Saaremaa/160 V | 50.0 | 17.9 | |
| DOBV 15/01 (Slovenia) | 245: DOBV Belgrade L41916 | 2 | Dobrava-Belgrade virus isolate HtSi_275_p2008; HtSi_1036_a2001 | 688 | 97.5 |
| 247: Saaremaa virus AJ616854 | 12 | HtSi_275_p2008; HtSi_1036_a2001 | 103 | 37.4 | |
| 254: Hantaan Q32 AB027097 | 76 | HtSi_275_p2008; HtSi_1036_a2001 | 69.8 | 24.2 | |
| 262: Soochong SC1 AY675349 | 100 | HtSi_275_p2008; HtSi_1036_a2001 | 51.8 | 24.4 | |
| DOBV 86/98 86/18 (Slovenia) | 245: DOBV Belgrade L41916 | 8 | Dobrava-Belgrade virus isolate HtSi_1012_a1997 | 159 | 43.1 |
| 246: DOBV Esl/81Aa/01 AY533120 | 5 | Dobrava-Belgrade virus isolate HtSi_1012_a1997 | 221 | 61.8 | |
| 247: Saaremaa virus AJ616854 | 9 | Dobrava-Belgrade virus isolate HtSi_1012_a1997 | 99.0 | 39.6 | |
See legend of Table 3 for details on presented results.
Table 5.
Detection and genetic characterization of Tula orthohantaviruses (TULV) from various endemic area in Europe: TULV isolates or laboratory strain were hybridized to the hantavirus sequences tiled on the PathogenID v3.0 resequencing microarray (seq222 to seq273; Table 1; Supplement 1).
| Tested virus (geographical origin) | Tiled sequence allowing detection (position on the chip) | BLASTN identification | Score (total) | Call rate (%) | |
|---|---|---|---|---|---|
| nbr° | sequences | ||||
| S666_13 Schlindermanderscheid (Germany) | 261:ProspectHill M34011 | 2 | S667_13_Marv; S666_13_Marv | 64.4 | 25.9 |
| 264: Kosice144/Ma/95 | 2 | S667_13_Marv; S666_13_Marv nucleocapsid protein gene, partial | 46.4 | 23.2 | |
| 265: Moravia5293Ma/94 | 2 | S667_13_Marv; S666_13_Marv nucleocapsid protein gene, partial | 107 | 38.1 | |
| TULV09_1000 Cunnersdorf (Germany) | 265: Moravia5293Ma/94 | 9 | 09_1000_Marv nucleocapsid protein gene, partial cds | 42.8 | 38.4 |
| TULV laboratory strain Moravia | 234: Perpignan1999 | 8 | Tula virus S segment nucleocapsid protein mRNA, complete cds | 55.4 | 23.8 |
| 244: CovictCreek107 | 49 | Tula virus segment S, strain Tula/Moravia/5294 Ma/94, genomic RNA | 42.8 | 16.2 | |
| 261: Prospect Hill M34011 | 7 | Tula/Moravia/5294 Ma/94, genomic RNA | 48.2 | 30.9 | |
| 263: Sennickerode_Sen05/204 | 72 | Tula/Moravia/5294 Ma/94, genomic RNA | 42.8 | 25.4 | |
| 264: Kosice144/Ma/95 | 10 | Tula/Moravia/5294 Ma/94, genomic RNA | 55.4 | 21.2 | |
| 265: Moravia5293Ma/94 | 1 | Tula virus segment S, strain Tula/Moravia/5302 Ma/94 | 688 | 97.5 | |
| 266: MG23/Omsk | 8 | Tula/Moravia/5302 Ma/94, genomic RNA | 44.6 | 21.4 | |
| TULV (Slovenia) | 265: Moravia5293Ma/94 | 1 | Tula virus isolate HtSi_1087_a1999 nucleocapsid protein gene, partial cds | 123 | 37.4 |
See legend of Table 3 for details on presented results.
Table 6.
Detection and genetic characterization of Seoul orthohantaviruses (SEOV) from various endemic area in Europe: SEOV isolates were hybridized to the hantavirus sequences tiled on the PathogenID v3.0 resequencing microarray (seq222 to seq273; Table 1; Supplement 1).
| Tested virus (geographical origin) | Tiled sequence allowing detection (position on the chip) | BLASTN identification\ | Score (total) | Call rate (%) | |
|---|---|---|---|---|---|
| nbr° | sequences | ||||
| Seoul Lyon 892 (France) | 255: Hantavirus SEOV Z37 | 1 | Seoul virus isolate LYON/Rn/FRA/2013/LYO852 segment S, complete | 235 | 64.0 |
| 257: Hantavirus SEOV L99 | 28 | Seoul virus isolate LYON/Rn/FRA/2013/LYO852 segment S, complete | 127 | 44.6 | |
| 258: SEOV 8039 | 10 | Seoul virus isolate LYON/Rn/FRA/2013/LYO852 segment S, complete | 140 | 53.6 | |
| Seoul Cherwell (UK) | 255: Hantavirus SEOV Z37 | 1 | Seoul virus nucleocapsid protein gene, complete cds Cherwell | 161 | 52.9 |
| 257: Hantavirus SEOV L99 | 1 | Seoul virus nucleocapsid protein gene, complete cds Cherwell | 91.5 | 48.1 | |
| 258: SEOV 8039 | 8 | Seoul virus nucleocapsid protein gene, complete cds Cherwell | 136 | 51.6 | |
See legend of Table 3 for details on presented results.
Discussion
Hantaviruses are zoonotic agents distributed world-wide. In Europe, rodent-borne hantaviruses are regularly provoking episodes of Hemorrhagic Fever with Renal Syndrome (HFRS). Tools to survey hantavirus circulation, geographical distribution and genetic features in the animal reservoir are essential to a better understanding and prevention of hantavirus infection in humans. Resequencing microarray has been shown powerful to precisely identify new genetic variants of emerging viruses15–21.
The present work represents a significant improvement of the resequencing microarray PathogenID developed through a collaborative study for detection and identification of orthohantaviruses circulating in Europe. The 1st and 2nd generations of PathogenID allowed to detect different viruses associated with hemorrhagic fevers, including hantaviruses; however they also showed their limits for the detection of PUUV20 which is the most common and widespread European hantavirus causing a mild form of HFRS, Nephropatia Emidemica (NE)2–8. Therefore, we have switched the strategy for the design of the 3rd generation PathogenID v3.0 used in this study. Tiled sequences did not target anymore the most conserved region of the genome, the L segment24, but the S segment encoding the N protein, which present two advantages: it is more efficient to discriminate variants within a species; more sequences are present in GenBank. After a critical analysis of the taxonomy, 52 representative hantavirus sequences were selected among those available at the time of the conception of the chip (Table 1). The resequencing methodology (Supplement 2) allowed to recognize both known viruses and previously unknown geographical variants.
Validation carried out by using hantavirus N protein encoding plasmids allowed to show a global tolerated divergence (up to 20%) between the tiled and the tested sequences for a correct identification of the PUUV prototype strain Sotkamo (Table 1, Table 2, Fig. 1; Supplement 3). A synergic effect in detection was observed when three viral species (PUUV; TULV; HTNV) were simultaneously tested (Supplement 4), most likely due their cross-contribution in hybridizing orthohantavirus conserved nucleotides, favoring identification by BLASTN analysis.
Identification of European hantaviruses, PUUV, DOBV, TULV, SEOV, TOPV present in tissue samples or in supernatants of Vero E6 cells infected with laboratory strains was demonstrated not only using the homologous sequences tiled on the chip, but also using phylogenetically distant sequences (Tables 3–7; Fig. 3). The key factor for the precise characterization of the tested hantavirus sequence was obviously its genetic distance from the tiled one (Fig. 3) outlining the importance of designing the chip from sequences encompassing the global diversity of hantaviruses. However, even for samples with a low Call Rate (CR: % of determined/total number of nucleotides following hybridization), precise taxonomical identification was possible when short specific fragments of significant sequence (about 15 nucleotides) were obtained for BLASTN analysis (Supplement 5). This was in particular the case for TOPV and PUUV Konnevesi variant in the present study (Tables 3–7). A similar observation was previously reported for hemorrhagic fever viruses detected by PathogenID v.220. These short stretches of highly conserved sequences among hantavirus could serve for developing other hybridization methods such as hybrid captures43.
Geographical variants of PUUV, the most frequent hantavirus in Europe, was correctly determined in the different endemic areas, such as France, Germany, Finland, and Slovenia (Table 3, Fig. 3). SEOV isolate, recently pointed out to circulate both in France and in UK39–41, was also precisely characterized despite neither Lyon nor Cherwell isolates, respectively, were known when the microarray was designed (Table 6). Interestingly, both isolates were detected and correctly identified from two heterologous sequences tiled on the microarray, SEOV and HTNV (Table 6). In any cases, when a tiled sequence did not achieve the determination of the exact geographical variant, it was at least designing the phylogenetic clade and the number of sequences tiled on the microarray always allowed to reach the deepest level of precision (Fig. 3).
Altogether, the results obtained with DOBV, TULV, SEOV and TOPV clearly outline the potential of Pathogen ID v3.0 to largely explore the hantavirus genetic space and to deliver precise identification of the species and local variants present in the infected tissue or in the cell supernatant. The suitability of this approach was demonstrated to map the wide diversity of hantaviruses within the European continent, including new variants unknown at the moment of the design of the chip. Detection by resequencing microarray which is applicable to both animal and human samples, is of interest for both research and public health aspects. Our results are promising to enlarge evaluation to other hantaviruses from different continents, both the pathogenic ones circulating in other endemic areas, such as Americas where they provoke severe HCPS and also in other animal reservoirs such as insectivores and bats.
Supplementary information
Acknowledgements
This work is the follow-up of the Programme Transversale de Recherche (PTR) ‘Detection of Emerging Viral Agents’ (DEVA) n° 246 at Institut Pasteur. The following European projects contributed to fund the research, both for material and fellowships: EMPERIE - European Management Platform for Emerging and Re-emerging Infectious Diseases Entities; EDEN - Emerging Diseases in a changing EuropeaN environment; EDENext (FP7-261504) - Biology and control of vector-borne infections in Europe and ANTIGONE - ANTIcipating the Global Onset of Novel Epidemics (FP7- 278976). The study was also supported by the German Federal Ministry of Education and Research (BMBF) for the “RoBoPub” consortium awarded to RGU (Grant 01KI1721A). We thank Nicolas Berthet, Mathilde Couteaudier, Stephan Drews, Sandra Lacote, Jean-Baptiste Pons, Jean-Michel Thiberge for their collaboration during the study.
Author Contributions
C.F., P.M., A.G., N.T. conceived the study. C.F. performed chip experiments, analysed and interpreted the data. S.M. performed RNA extraction experiments. M.E. was involved in experiments with plasmids. G.C. performed the phylogenetic analysis. P.M., S.M., F.S., T.A.-Z., M.K., T.S., O.V., L.M., R.G.U., M.G. were involved in providing viruses from endemic area. N.T., A.G., S.V.W. supported the work. V.C. and J.C.M. helped in using microarray platform. C.F. and N.T. drafted the manuscript. P.M., G.C., A.G. and R.G.U. revised the manuscript. All authors approved the final version of the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-47508-7.
References
- 1.International Committee on Taxonomy of Viruses (ICTV). Virus Taxonomy: Release, https://talk.ictvonline.org/taxonomy/ (2018).
- 2.Reusken C, Heyman P. Factors driving hantavirus emergence in Europe. Curr. Opin. Virol. 2013;3:92–99. doi: 10.1016/j.coviro.2013.01.002. [DOI] [PubMed] [Google Scholar]
- 3.Vapalahti O, et al. Hantavirus infections in Europe. Lancet Infect. Dis. 2003;3:653–661. doi: 10.1016/S1473-3099(03)00774-6. [DOI] [PubMed] [Google Scholar]
- 4.Olsson GE, Leirs H, Henttonen H. Hantaviruses and their hosts in Europe: reservoirs here and there, but not everywhere? Vector Borne Zoonotic Dis. 2010;10:549–561. doi: 10.1089/vbz.2009.0138.. [DOI] [PubMed] [Google Scholar]
- 5.Heyman P, Vaheri A, Lundkvist A, Avsic-Zupanc T. Hantavirus infections in Europe: from virus carriers to a major public-health problem. Expert Rev. Anti. Infect. Ther. 2009;7:205–217. doi: 10.1586/14787210.7.2.205.. [DOI] [PubMed] [Google Scholar]
- 6.Vaheri A, et al. Hantavirus infections in Europe and their impact on public health. Rev. Med. Virol. 2013;23:35–49. doi: 10.1002/rmv.1722. [DOI] [PubMed] [Google Scholar]
- 7.Viel JF, et al. Environmental risk factors for haemorrhagic fever with renal syndrome in a French new epidemic area. Epidemiol. Infect. 2011;139:867–874. doi: 10.1017/S0950268810002062.. [DOI] [PubMed] [Google Scholar]
- 8.Klempa B, Radosa L, Kruger DH. The broad spectrum of hantaviruses and their hosts in Central Europe. Acta Virol. 2013;57:130–137. doi: 10.4149/av_2013_02_130. [DOI] [PubMed] [Google Scholar]
- 9.Watson DC, et al. Epidemiology of Hantavirus infections in humans: a comprehensive, global overview. Crit. Rev. Microbiol. 2014;40:261–272. doi: 10.3109/1040841X.2013.783555.. [DOI] [PubMed] [Google Scholar]
- 10.Kruger DH, Figueiredo LT, Song JW, Klempa B. Hantaviruses–globally emerging pathogens. J. Clin. Virol. 2015;64:128–136. doi: 10.1016/j.jcv.2014.08.033.. [DOI] [PubMed] [Google Scholar]
- 11.Zeier M, et al. New ecological aspects of hantavirus infection: a change of a paradigm and a challenge of prevention–a review. Virus Genes. 2005;30:157–180. doi: 10.1007/s11262-004-5625-2. [DOI] [PubMed] [Google Scholar]
- 12.Guo WP, et al. Phylogeny and origins of hantaviruses harbored by bats, insectivores, and rodents. PLoS Pathog. 2013;9:e1003159. doi: 10.1371/journal.ppat.1003159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heyman P, Thoma BR, Marié JL, Cochez C, Essbauer SS. In Search for Factors that Drive Hantavirus Epidemics. Front. Physiol. 2012;3:237. doi: 10.3389/fphys.2012.00237.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jonsson CB, Figueiredo LT, Vapalahti O. A global perspective on hantavirus ecology, epidemiology, and disease. Clin. Microbiol. Rev. 2010;23:412–441. doi: 10.1128/CMR.00062-09.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nordström H, et al. Microarray technology for identification and distinction of hantaviruses. J. Med. Virol. 2004;72:646–655. doi: 10.1002/jmv.20041. [DOI] [PubMed] [Google Scholar]
- 16.Kothiyal, P. et al. An overview of custom array sequencing. Curr. Protoc. Hum. Genet. Chapter 7, Unit 7.17, 10.1002/0471142905.hg0717s61 (2009). [DOI] [PMC free article] [PubMed]
- 17.Berthet N, et al. Massively parallel pathogen identification using high-density microarrays. Microb. Biotechnol. 2008;1:79–86. doi: 10.1111/j.1751-7915.2007.00012.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berthet N, et al. Maculopapular lesions in the Central African Republic. Lancet. 2011;378:1354. doi: 10.1016/S0140-6736(11)61142-2. [DOI] [PubMed] [Google Scholar]
- 19.Berthet N, et al. High-density resequencing DNA microarrays in public health emergencies. Nat. Biotechnol. 2010;28:25–27. doi: 10.1038/nbt0110-25.. [DOI] [PubMed] [Google Scholar]
- 20.Filippone C, et al. Molecular diagnostic and genetic characterization of highly pathogenic viruses: application during Crimean-Congo haemorrhagic fever virus outbreaks in Eastern Europe and the Middle East. Clin. Microbiol. Infect. 2013;19:E118–128. doi: 10.1111/1469-0691.12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dacheux L, et al. Application of broad-spectrum resequencing microarray for genotyping rhabdoviruses. J. Virol. 2010;84:9557–9574. doi: 10.1128/JVI.00771-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Firth C, et al. Detection of zoonotic pathogens and characterization of novel viruses carried by commensal Rattus norvegicus in New York City. MBio. 2014;5:e01933–14. doi: 10.1128/mBio.01933-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sachsenröder J, et al. Metagenomic identification of novel enteric viruses in urban wild rats and genome characterization of a group A rotavirus. J. Gen. Virol. 2014;95:2734–2747. doi: 10.1099/vir.0.070029-0.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Plyusnin, A. & Elliott, R. M. Bunyaviridae: molecular and cellular biology. (Norfolk, UK: Caister Academic Press, 2011).
- 25.Klempa B, et al. Complex evolution and epidemiology of Dobrava-Belgrade hantavirus: definition of genotypes and their characteristics. Arch. Virol. 2013;158:521–529. doi: 10.1007/s00705-012-1514-5.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Temonen M, et al. Susceptibility of human cells to Puumala virus infection. J. Gen. Virol. 1993;74:515–8. doi: 10.1099/0022-1317-74-3-515. [DOI] [PubMed] [Google Scholar]
- 27.Vapalahti O, et al. Isolation and characterization of Tula virus, a distinct serotype in the genus Hantavirus, family Bunyaviridae. J. Gen. Virol. 1996;77:3063–3067. doi: 10.1099/0022-1317-77-12-3063. [DOI] [PubMed] [Google Scholar]
- 28.Vapalahti O, et al. Isolation and characterization of a hantavirus from Lemmus sibiricus: evidence for host switch during hantavirus evolution. J. Virol. 1999;73:5586–5592. doi: 10.1128/jvi.73.7.5586-5592.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Plyusnina A, et al. Analysis of complete Puumala virus genome, Finland. Emerg. Infect. Dis. 2012;18:2070–2072. doi: 10.3201/eid1811.120747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Razzauti M, Plyusnina A, Henttonen H, Plyusnin A. Accumulation of point mutations and reassortment of genomic RNA segments are involved in the microevolution of Puumala hantavirus in a bank vole (Myodes glareolus) population. J. Gen. Virol. 2008;89:1649–1660. doi: 10.1099/vir.0.2008/001248-0. [DOI] [PubMed] [Google Scholar]
- 31.Castel G, et al. Complete Genome and Phylogeny of Puumala Hantavirus Isolates Circulating in France. Viruses. 2015;7:5476–5488. doi: 10.3390/v7102884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ettinger J, et al. Multiple synchronous outbreaks of Puumala virus, Germany, 2010. Emerg. Infect. Dis. 2012;18:1461–1464. doi: 10.3201/eid1809.111447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schlegel M, et al. Tula virus infections in the Eurasian water vole in Central Europe. Vector Borne Zoonotic Dis. 2012;12:503–513. doi: 10.1089/vbz.2011.0784.. [DOI] [PubMed] [Google Scholar]
- 34.Schmidt S, et al. High genetic structuring of Tula hantavirus. Arch. Virol. 2016;161:1135–1149. doi: 10.1007/s00705-016-2762-6.. [DOI] [PubMed] [Google Scholar]
- 35.Faber M, et al. Puumala virus outbreak in Western Thuringia, Germany, 2010: epidemiology and strain identification. Zoonoses Public Health. 2013;60:549–554. doi: 10.1111/zph.12037. [DOI] [PubMed] [Google Scholar]
- 36.Schmidt - Chanasit J, et al. Extensive host sharing of central European Tula virus. J. Virol. 2010;84:459–474. doi: 10.1128/JVI.01226-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Korva M, et al. Phylogeographic diversity of pathogenic and non-pathogenic hantaviruses in Slovenia. Viruses. 2013;5:3071–3087. doi: 10.3390/v5123071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Korva M, Duh D, Puterle A, Trilar T, Zupanc TA. First molecular evidence of Tula hantavirus in Microtus voles in Slovenia. Virus Res. 2009;144:318–322. doi: 10.1016/j.virusres.2009.04.021. [DOI] [PubMed] [Google Scholar]
- 39.Dupinay T, et al. Detection and genetic characterization of Seoul virus from commensal brown rats in France. Virol. J. 2014;11:32. doi: 10.1186/1743-422X-11-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McElhinney LM, et al. High prevalence of Seoul hantavirus in a breeding colony of pet rats. Epidemiol. Infect. 2017;145:3115–3124. doi: 10.1017/S0950268817001819.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jameson LJ, et al. The continued emergence of hantaviruses: isolation of a Seoul virus implicated in human disease, United Kingdom, October 2012. Euro Surveill. 2013;18:4–7. [PubMed] [Google Scholar]
- 42.Lefort V, Longueville JE, Gascuel O. SMS: Smart Model Selection in PhyML. Mol. Biol. Evol. 2017;34:2422–2424. doi: 10.1093/molbev/msx149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gaudin M, Desnues C. Hybrid Capture-Based Next Generation Sequencing and Its Application to Human Infectious Diseases. Front. Microbiol. 2018;9:2924. doi: 10.3389/fmicb.2018.02924. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.