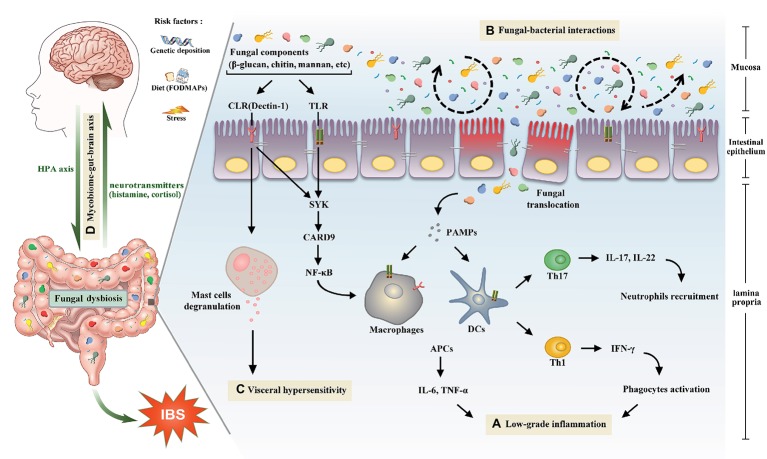
Figure 1.
The potential role of gut mycobiome in the pathogenesis of irritable bowel syndrome. Gut mycobiome is involved in the existing mechanisms of irritable bowel syndrome including low-grade inflammation, intestinal dysbiosis, visceral hypersensitivity, and brain-gut interactions. (A) Pattern recognition receptors of human immune system, including CLRs and TLRs, can recognize fungal components such as β-glucan, chitin, and mannan, which will drive a series of downstream signaling pathways, then cause proinflammatory reactions and lead to low-grade inflammation in the intestine. (B) Tight interactions between fungi and bacteria are observed in the intestinal microecosystem. Fluctuations of either of the two species will lead to the intestinal dysbiosis. (C) Recognition of β-glucan by CLRs can trigger mucosal mast cells degranulation, which is a crucial mechanism of visceral hypersensitivity. (D) Intestinal mycobiome is another essential member of brain-gut axis aside from bacteria. Central nervous system can influence the composition of gut mycobiome through the HPA axis, which is closely associated with cognitive functions and gastrointestinal dynamics and colonic hypersensitivity. In turn, the mycobiome can release neuromediators such as cortisol and histamine to react on the central nervous system. Abbreviations: CLRs, C-type lectin receptors; TLRs, Toll-like receptors; SYK, spleen tyrosine kinase; CARD9, caspase recruitment domain family, member 9; NF-κB, nuclear factor-kappa B; HPA, hypothalamic-pituitary-adrenal; PAMPs, pathogen-associated molecular patterns; DCs, dendritic cells; APCs, antigen presenting cells; IL, interleukin; TNF, tumor necrosis factor; Th, T helper cell; IFN-γ, interferon gamma.