Figure 1.
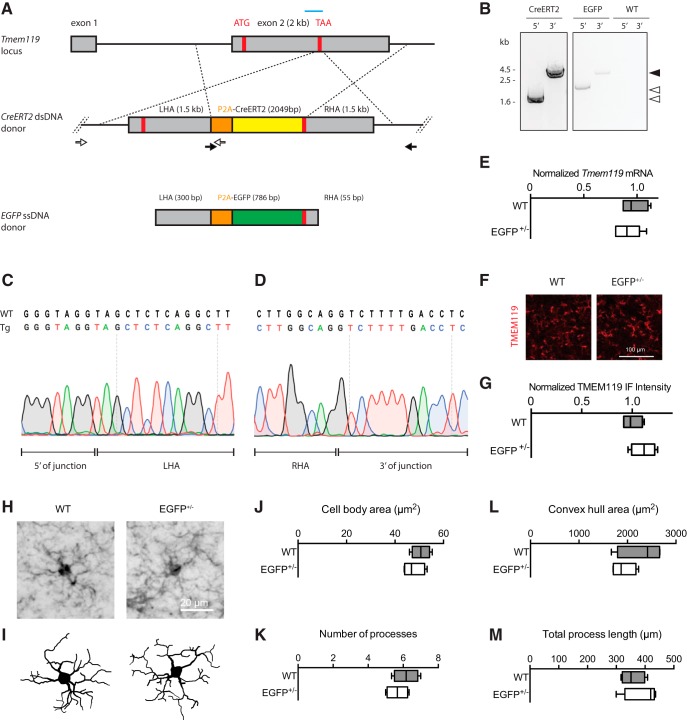
Generation and validation of Tmem119-EGFP and Tmem119-CreERT2 knock-in lines. A, Experimental strategy using a guide RNA (blue bar) to introduce a double-strand break at the Tmem119 stop codon in mouse zygotes and an injection mixture containing EGFP ssDNA template with short homology arms of 55–300 bp or CreERT2 dsDNA template with long 1.5 kb homology arms. Each knock-in is designed to cause in-frame insertion of P2A peptide followed by EGFP or CreERT2 and a stop codon. Not drawn to scale. B, Confirmation of insertion by amplification of fragments spanning the 5′ and 3′ junctions using sets of primers binding inside the template and outside the homology arms (A, open and closed arrows with arrowheads indicating corresponding products). C, D, Sanger sequencing across the 5′ and 3′ junction of the CreERT2 line and comparison to WT sequence indicate seamless insertion in the transgenic mice (Tg). E, qPCR analysis of gene expression for Tmem119 in WT controls and Tmem119-EGFP+/− knock-in animals. N = 4 WT, N = 6 Tmem119-EGFP+/− mice. F, Representative TMEM119 immunoreactivity in WT controls and Tmem119-EGFP+/− knock-in animals. G, Relative intensity of TMEM119 immunostaining signal. N = 4 WT, N = 4 Tmem119-EGFP+/− mice. H, I, Representative IBA-immunostained microglia and corresponding Neurolucida traces in WT and Tmem119-EGFP+/− knock-in mice. J–M, Quantified cell body area, number of processes, convex hull area, and total process length of microglia in WT and Tmem119-EGFP+/− knock-in mice. N = 12 microglia from four mice per genotype. TAA (red bar), stop codon.