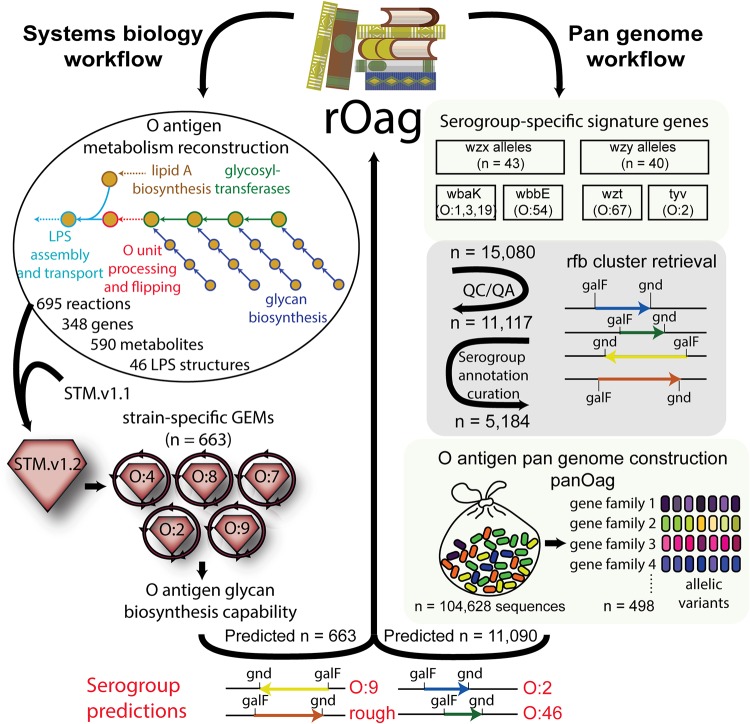
FIG 3.
O-antigen gene island database construction and modeling workflow chart. O-antigen gene islands were retrieved from the PATRIC database and quality checked (48). An O-antigen pangenome was constructed by pooling together the O-antigen island gene content across strains. Genes were clustered into gene families by means of sequence homology using CD-HIT (45), and reference gene clusters were chosen for all 46 serogroups. Genes were then annotated with a metabolic function and used to reconstruct an O-antigen metabolic network. Serogroup annotations were derived from the metadata associated with each genome, and serogroup predictions were made using serogroup-specific signature orthologous groups.