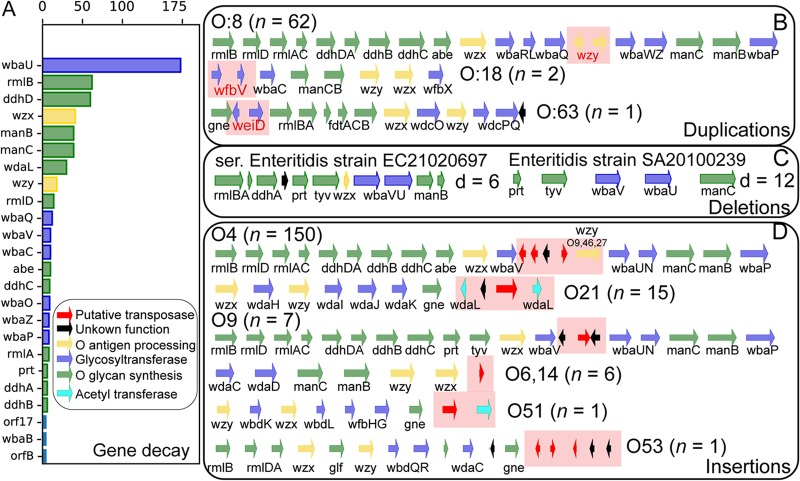
FIG 4.
Sources of genetic variability in the content of rfb clusters. (A) Gene decay across orthologous groups. (B) Tandem gene duplications. (C) Gene deletions (d = no. genes missing with respect to the reference rfb cluster). In these cases, the strains likely exhibit a rough phenotype (e.g., they do not synthesize O-antigens). (D) Gene insertions. Note that only one sample of each serogroup affected by insertions is shown but that the rfb cluster may vary in content across the n strains (due to additional insertions, deletions, or duplications). Similarly, we only show a subset of serogroups and rfb clusters with gene duplication and deletion events because there are too many serogroups which are affected by these phenomena (see Table S7 for a full list).