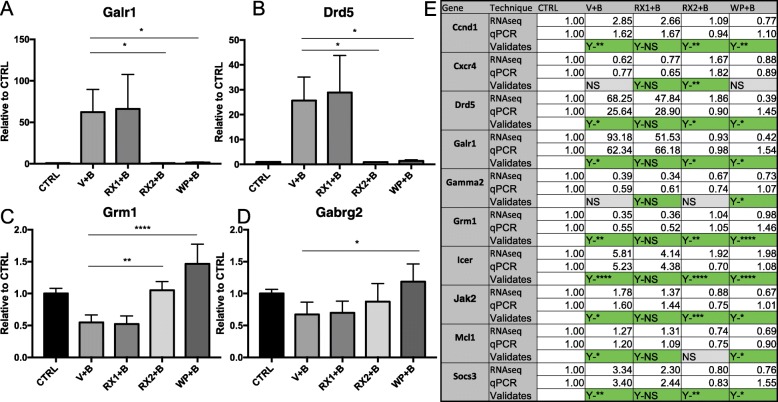
Fig. 6.
qRT-PCR validation of BDNF regulated genes in presence of JAK/STAT inhibitors. Primary neurons 9 DIV were pretreated with100nM Ruxolitinib (RX1), 10uM Ruxo (RX2), 10uM WP1066 (WP) or DMSO for 1 h before the addition of BDNF. a-d Graphical representation of selected results of real time PCR analysis using Taqman probe and primers. RNA was extracted from cells collected 4 h after BDNF administration. mRNA levels are shown as the mean values (±SEM) of the ratio relative to DMSO+Water control group for a Galr1 n = 3, b Drd5 n = 3, c Grm1 n = 4, d Gabrg2 n = 4. e Compilation of all genes tested for validation with qRT-PCR represented alphabetically. Values are the ratio relative to DMSO+Water for RNAseq (top row) and qRT-PCR (2nd row) for comparison. Indication of a validating result (3rd row) is marked in green. Results marked with Yes(Y) followed by the level of significance indicate a significant result consistent with RNA-seq. Yes- Not significant (Y-NS) seen in all the RX1 is the expected result consistent with our RNA-seq findings. Measurements that did not reach significance are marked in gray as Not Significant (NS). Significance was tested using One-Way ANOVA followed by Tukey’s test for multiple comparisons. (*p < 0.05, **p < 0.01, ***p < 0.0005, ****p < .0001)