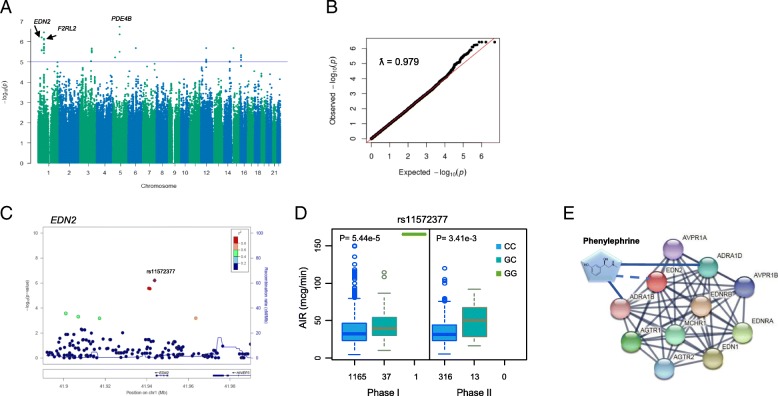
Fig. 4.
Meta-analysis and subsequent analyses on top hits. a Manhattan plot and b Q-Q plot of the meta-analysis for phenylephrine average infusion rate. Top loci with P < 10–6 were labeled. Genome inflation factor ƛ was 0.979. c Regional association for EDN2 in meta-analyses for phenylephrine infusion rate. 800 kb flanking the genomic region of the lead SNP, marked as purple diamond, were illustrated. d Boxplot of average infusion rate against the rs11572377 genotypes in phase I and phase II cohorts. Raw P value refers to allelic association under additive model of linear regression adjusted for the corresponding covariates. e Protein-protein interaction network for EDN2 identified by STRING. The nodes and edges respectively represent encoded proteins and evidence-based functional interaction derived from a combined score which was computed by combining the probabilities from up to 7 different resources and corrected for the probability of randomly observing an interaction. Only high confidence interactions were shown here (interaction score ≥ 0.9). Phenylephrine node was superimposed to the existing interactive plot. A solid line was created between phenylephrine and ADRA1B or phenylephrine and ADRA1D because of converging evidence from literature. A dashed line was created to show the hypothetical connect between phenylephrine and EDN2 due to lack of solid evidence from literature