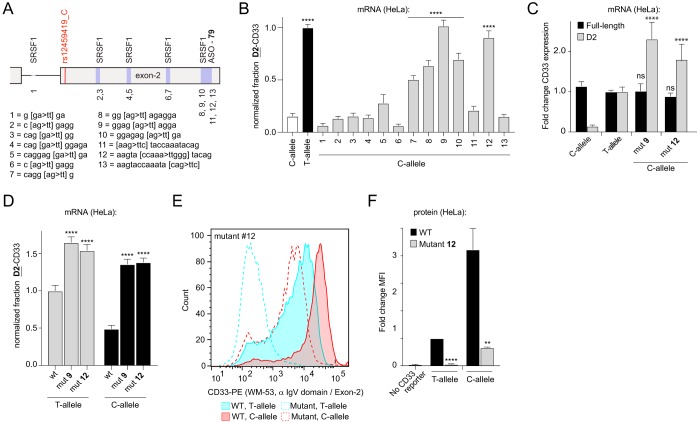
FIG 5.
Disrupting a predicted SRSF1 binding site near the 3′ end of exon 2 increases CD33 exon 2 skipping. (A) Overview of the predicted SRSF1 binding sites, as well as the ASO targeting sequence in CD33 exon 2. The numbered key indicates the identity of binding/targeting site mutants used in panel B. (B and D) T-allele-normalized fraction of CD33 mRNA transcripts lacking exon 2, as measured by RT-qPCR in HeLa cells transfected with CD33 minigenes carrying mutations as shown in panel A. (C) Fold change of full-length CD33 and D2-CD33 mRNA transcripts in HeLa cells overexpressing a mutant CD33 minigene compared to WT CD33 minigene-overexpressing cells, as measured by RT-qPCR. (E and F) Flow cytometry analysis of CD33 surface levels using a labeled antibody targeting CD33 C2 domain in HeLa cells overexpressing WT or mutant 12 containing CD33 minigenes in combination with either rs12459419C or rs12459419T. The MFIs were normalized to cells transfected with WT-CD33 minigenes carrying rs12459419T. Error bars indicate means plus the SEM. A one-way ANOVA (B to D) and a two-tailed t test (F) were performed. **, P ≤ 0.01; ****, P ≤ 0.0001; ns, not significant. n = 3 biological replicates for each plot.