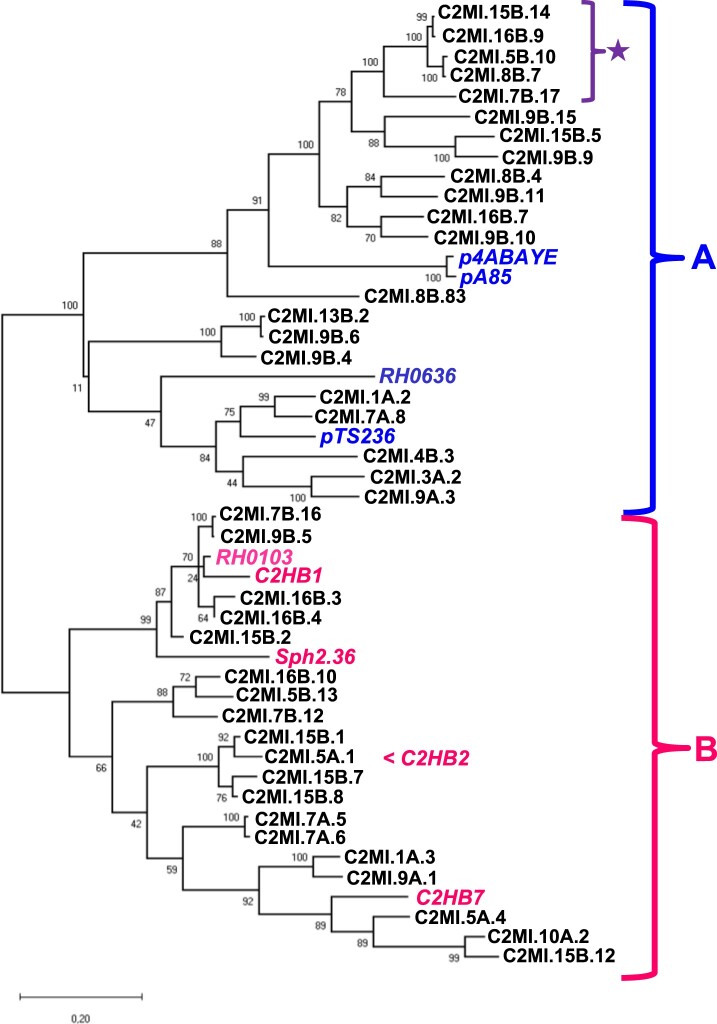
Figure 2.
Phylogenetic tree of the nucleotide sequences of representatives of each BMMF2 isolate in comparison to the plasmids of A. baumanii p4ABAYE, pA85-1 and pTS236, genomic isolates from rat gut pRGRH0103 (RH103) and pRGRH0636 (RH0636) in clade A, as well as Sphinx2.36 and our previous BMMF1 isolates C2MI1 and C2HB7 in clade B. The recently isolated C2MI.5A.1 represents the full-length genome of C2HB2. The star indicate BMMF2 isolates of which the nucleotide sequence is almost identical in the overlapping nucleotide sequence of the respective “small” BMMF2 isolate. The evolutionary history was inferred by using the Maximum Likelihood method and Tamura-Nei model [38]. The bootstrap consensus tree inferred from 500 replicates [29] is taken to represent the evolutionary history of the taxa analysed [29]. Branches corresponding to partitions reproduced in less than 50% bootstrap replicates are collapsed. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (500 replicates) are shown next to the branches [29]. Initial tree(s) for the heuristic search were obtained by applying the Neighbour-Joining method to a matrix of pairwise distances estimated using the Maximum Composite Likelihood (MCL) approach. This analysis involved 48 nucleotide sequences. There were a total of 3170 positions in the final dataset. Evolutionary analyses were conducted in MEGA X [26].