Figure 1.
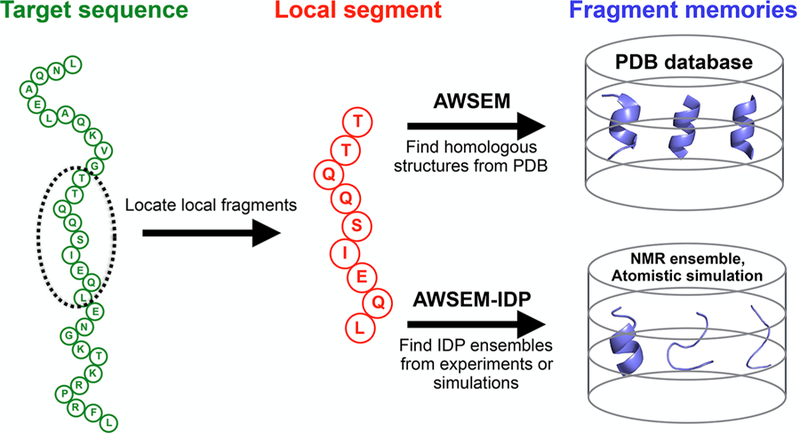
Schematic diagram of the fragment memory terms in AWSEM and AWSEM-IDP. In both AWSEM and AWSEM-IDP, the target sequence (green) is assigned into short local segments (red). Then structural fragments called “memories” (blue) are chosen to bias the local segment. The original forms of AWSEM search for fragment memories from the PDB database, while AWSEM-IDP utilizes NMR ensembles or atomistic simulation trajectories to construct the fragment library. The example sequence shown here is the amino-terminal domain of phage 434 repressor (PDB ID: 1R69).
