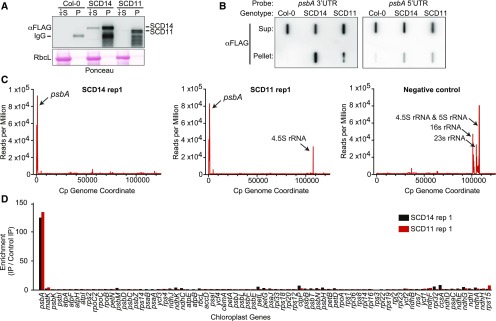
Figure 2.
RIP-Seq Analysis of RNAs That Coimmunoprecipitate with SCD14 and SCD11.
(A) Immunoprecipitation of SCD14 and SCD11. Stromal extracts from transgenic Arabidopsis expressing the indicated protein or from the Col-0 progenitor were used for immunoprecipitation with anti-FLAG antibody. The pellet (P) and supernatant (S) fractions were analyzed by immunoblot analysis using anti-FLAG antibody. An excerpt of the Ponceau S–stained filter is shown to illustrate the abundance of the large subunit of Rubisco (RbcL), which serves as a loading control. An equal proportion of each pellet fraction was analyzed; one-fourth that proportion of each supernatant was analyzed to avoid overloading the lane.
(B) Slot-blot hybridization analysis of RNAs that coimmunoprecipitate with SCD14 and SCD11. RNA was extracted from the immunoprecipitations analyzed in (A) and applied to nylon membrane via a slot-blot manifold. The same proportion of all samples was analyzed to illustrate the partitioning of psbA RNA between the pellet and supernatant (Sup) fractions. Replicate blots were hybridized with oligonucleotide probes specific for the psbA 5ʹ UTR or 3ʹ UTR.
(C) Plastome-wide view of RNAs that coimmunoprecipitate with SCD11 and SCD14. Results are plotted as the sequence coverage in consecutive 10-nucleotide windows along the chloroplast genome (accession NC_000932.1) per million reads mapped to the chloroplast genome. The negative control used an antibody that does not detect proteins in Arabidopsis together with extract from the SCD14 line. Data for replicate experiments are shown in Supplemental Figure 2A. Read counts for all RIP-seq experiments are provided in Supplemental Data Set 1.
(D) Enrichment of RNA from each protein-coding chloroplast gene in SCD11 and SCD14 coimmunoprecipitates. The ratio of normalized reads/gene in the experimental versus control immunoprecipitations (IP) is shown for replicate 1. Data for replicate experiments are shown in Supplemental Figure 2A.