Figure 3.
Analysis of Off-Target Binding by SCD14 and SCD11.
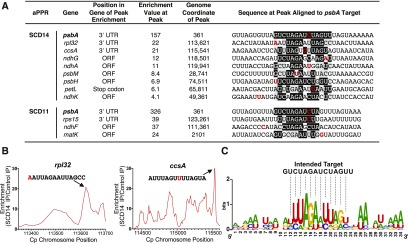
(A) Genes whose transcripts were enriched more than threefold in both replicate SCD14 and SCD11 coimmunoprecipitates. The position of peak enrichment within each gene (see [B] and Supplemental Figure 3) and the magnitude of enrichment at that peak (average of replicates) are indicated, together with sequence motifs within the peak that resemble the intended binding site for SCD14 and SCD11. Matches to the intended binding site in the psbA 3ʹ UTR are shaded in black. Transition mismatches are shaded in gray. The nucleotide at the peak of enrichment (see profiles in Figure 3B and Supplemental Figure 3) is marked in red. ORF, open reading frame.
(B) Local sequence enrichment profile for the off-target sites in rpl32 and ccsA. The regions shown span the open reading frame and UTRs of the indicated gene. The displayed sequences overlap the point of maximum enrichment (nucleotide at peak marked in red) and resemble the intended binding site of SCD14 (see [A]). Analogous plots for other genes listed in (A) are shown in Supplemental Figure 3. IP, immunoprecipitate.
(C) Sequence logo derived from the off-target sites of SCD14. Sequences listed in (A) were weighted according to the degree to which RNA from the corresponding region was enriched in the SCD14 immunoprecipitation and analyzed with WebLogo (Crooks et al., 2004).