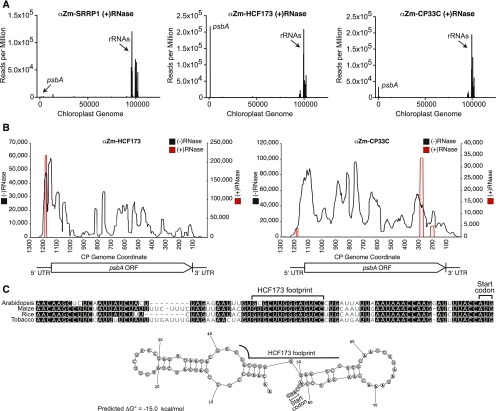
Figure 6.
High-Resolution RIP-Seq Analysis of HCF173 and CP33C Pinpointing Interaction Sites in the psbA mRNA.
Experiments were performed as in Figure 5 except that the stromal extract was briefly treated with RNase I prior to antibody addition.
(A) Average sequence coverage in consecutive 10-nucleotide windows along the chloroplast genome, per million reads mapped to the chloroplast genome (NCBI NC_001666).
(B) Sequence coverage along the psbA mRNA, with [red, (+) RNase] or without [black, (−) RNase] a pretreatment of stroma with RNase I. The number of reads per million mapped to the chloroplast genome (y axes) is shown according to position along the chloroplast genome (x axes). The sequences of the RNase-resistant peaks are shown in (C) (HCF173) and Supplemental Figure 4B (CP33C).
(C) Site of HCF173 interaction in the psbA 5ʹ UTR. The RNase-resistant sequence that coimmunoprecipitates with HCF173 (HCF173 footprint) is marked on a multiple sequence alignment of the psbA 5ʹ UTR from Arabidopsis, maize, tobacco (Nicotiana tabacum), and rice (Oryza sativa). A conserved secondary structure was predicted by Dynalign (Fu et al., 2014) using the maize and Arabidopsis sequences as input; the prediction for maize is shown below.