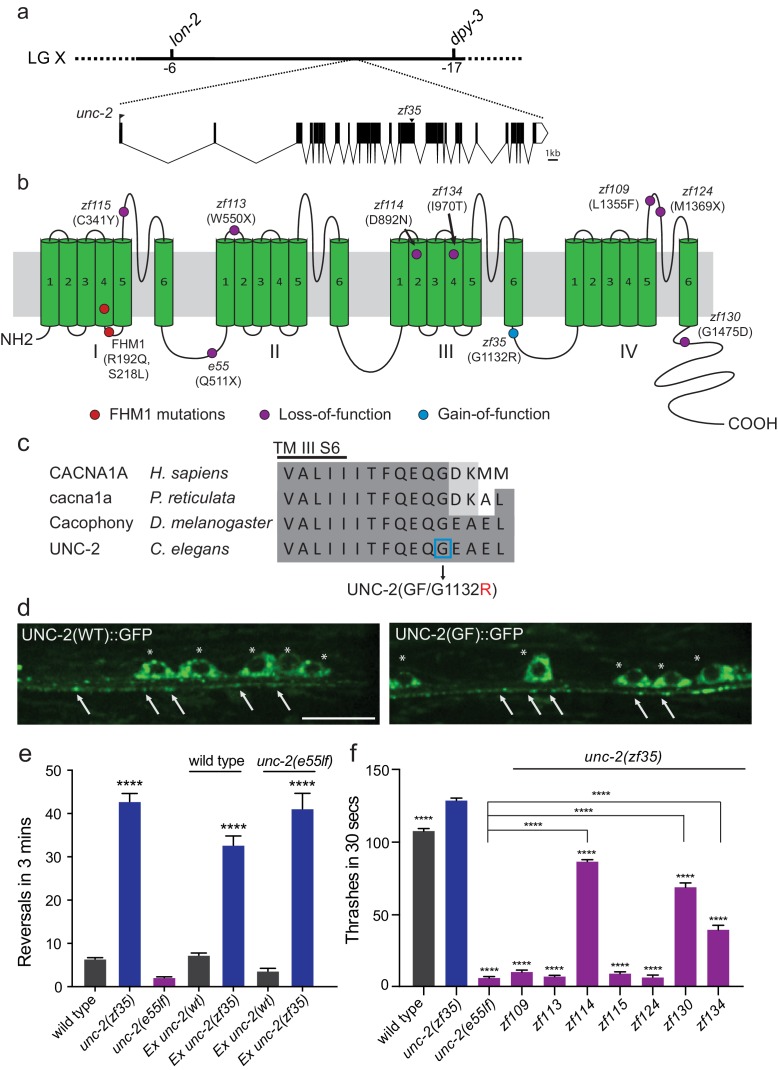
Figure 2. zf35 is a novel allele of the CaV2α subunit gene unc-2.
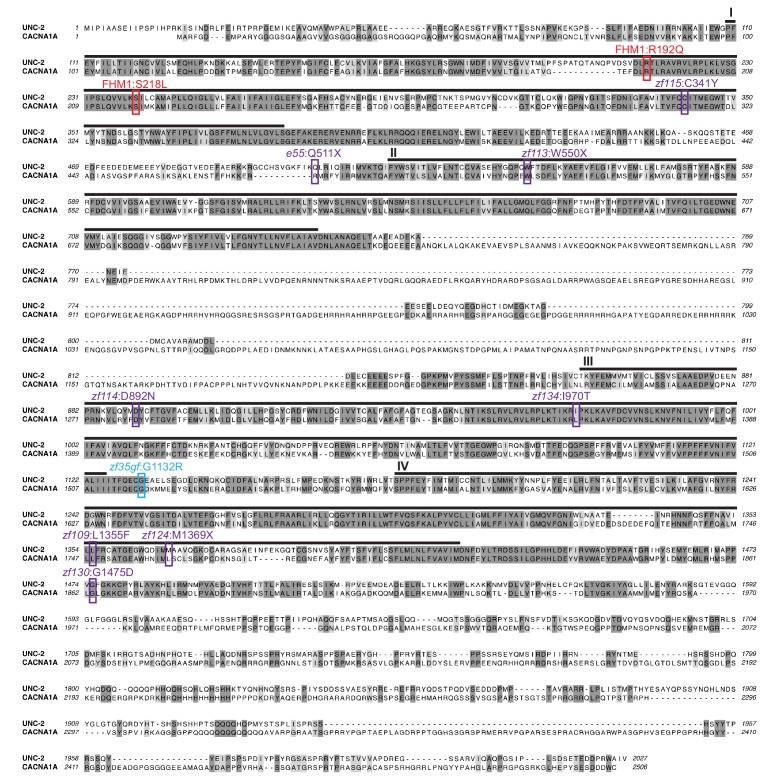
(a) The genetic map and gene structure of unc-2. Coding sequences are represented as black boxes. The zf35 allele is a single nucleotide transition (GGA to AGA) resulting in a glycine to arginine (G to R) amino acid substitution at position 1132. (b) Diagram of the secondary structure of UNC-2/CaV2α. UNC-2/CaV2α consists of four domains (I–IV) each containing six alpha-helix transmembrane (TM) segments (S1 – S6). The UNC-2 (G1132R) mutation localizes in the intracellular loop between TM domain III and IV, indicated by the blue circle. Purple circles indicate positions of intragenic unc-2(zf35) suppressors, red circles indicate the location of human FHM1 mutations. (c) The G1132R mutation occurs in a highly conserved region of the CaV2α subunit. Amino acid alignment of C-terminus region of the transmembrane III alpha-helix segment 6 (III S6) and the beginning of the third intracellular loop of CaV2α subunits from human (Homo sapiens, CACNA1A), rainbow fish (Poecilia reticulata, cacna1a), fly (Drosophila melanogaster, Cacophony) and nematode (C. elegans. UNC-2). Identities are shaded in dark gray, similarities in light gray. Location of the G1132R mutation is indicated. (d) Representative images of GFP tagged UNC-2(WT) and UNC-2(GF/G1132) in the ventral nerve cord. Asterisks point the cell bodies of the motor neurons and arrows indicate the presynaptic sites. Both constructs are expressed under pan-neuronal promoter tag-168. Scale bar, 10 μm. (e) Quantification of the reversal frequency: wild type (6.6 ± 0.4, n = 70), unc-2(zf35) (43.3 ± 1.9, n = 65), unc-2(e55lf) (2.4 ± 0.2, n = 59), wild-type animals expressing unc-2(wt) transgene (7.5 ± 0.6, n = 10) and unc-2(zf35) transgene (33 ± 2.1, n = 22), and unc-2(e55lf) rescued with unc-2(wt) transgene (3.8 ± 0.7, n = 12) and unc-2(zf35) transgene (41.3 ± 3.6, n = 21). Error bars represent SEM for at least three trials with indicated totaling animals number. Statistical difference from wild type ****p<0.0001, one-way ANOVA with Dunnett’s multiple comparisons test. (f) Intragenic unc-2(lf) mutations suppress unc-2(zf35) hyperactive locomotion. Shown are numbers of thrashes in 30 s in M9 for the wild type (107.0 ± 14.0, n = 60), unc-2(zf35) (128.1 ± 13.5, n = 60), unc-2(lf) (4.8 ± 2.1, n = 57), unc-2(zf35 zf109) (6.9 ± 4.3, n = 53); unc-2(zf35 zf113) (5.6 ± 3.7 thrashes, n = 57); unc-2(zf35 zf114) (80.2 ± 9.9, n = 60); unc-2(zf35 zf115) (6.9 ± 3.8, n = 56); unc-2(zf35 zf124) (5.3 ± 3.1, n = 57); unc-2(zf35 zf130) (67.1 ± 22.5, n = 58); unc-2(zf35 zf134) (31.2 ± 17.9, n = 50). Error bars represent SEM. Statistical difference from unc-2(zf35) mutants unless otherwise indicated, ****p<0.0001, one-way ANOVA with Tukey’s multiple comparisons test.