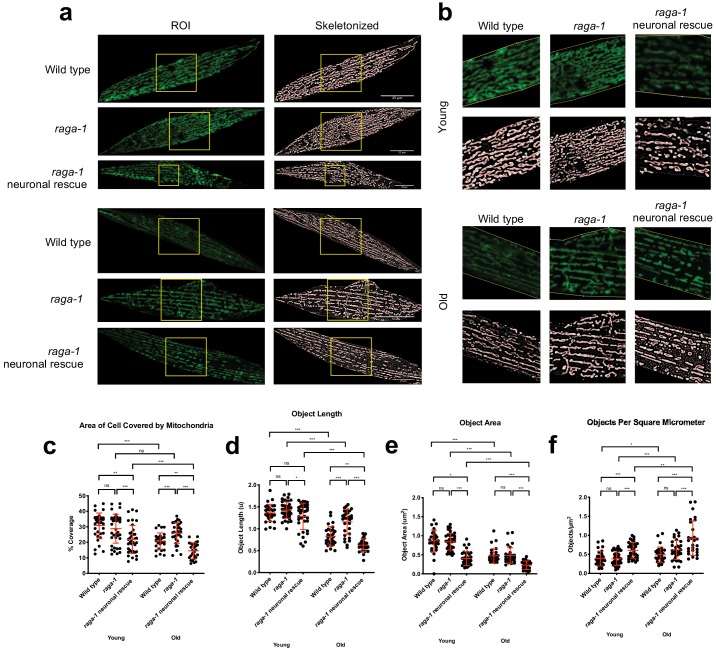
Figure 4. Neuronal RAGA-1 drives mitochondrial fragmentation in muscle cells.
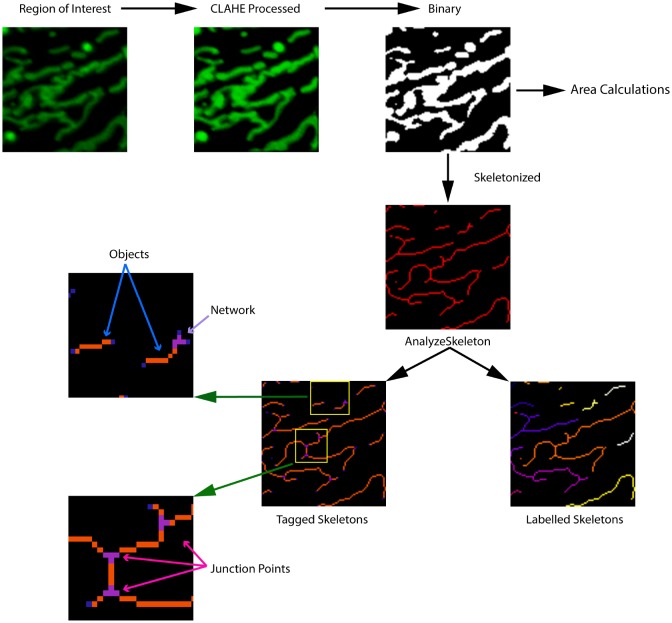
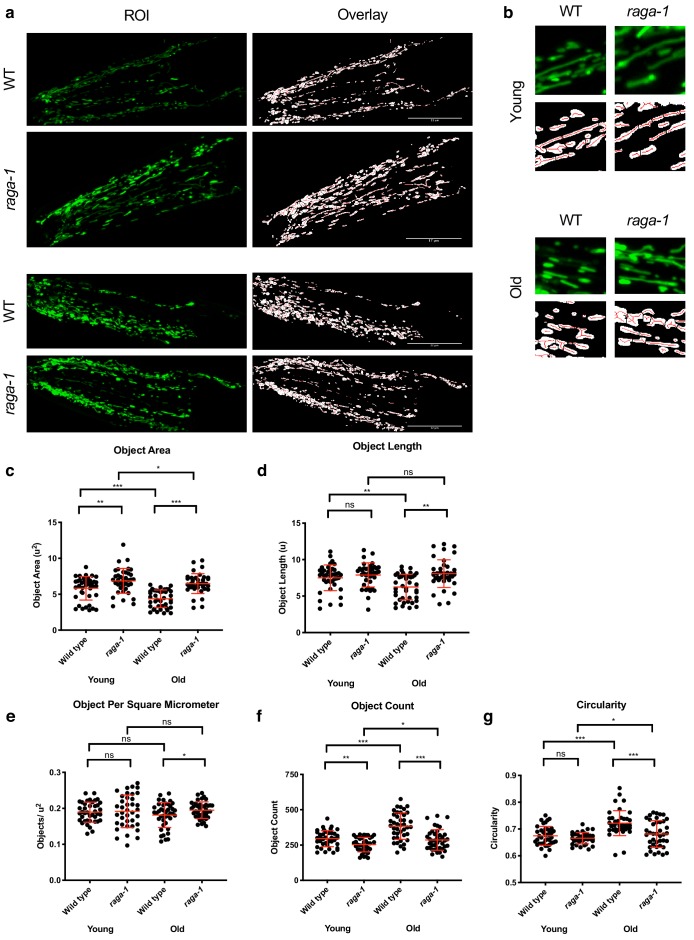
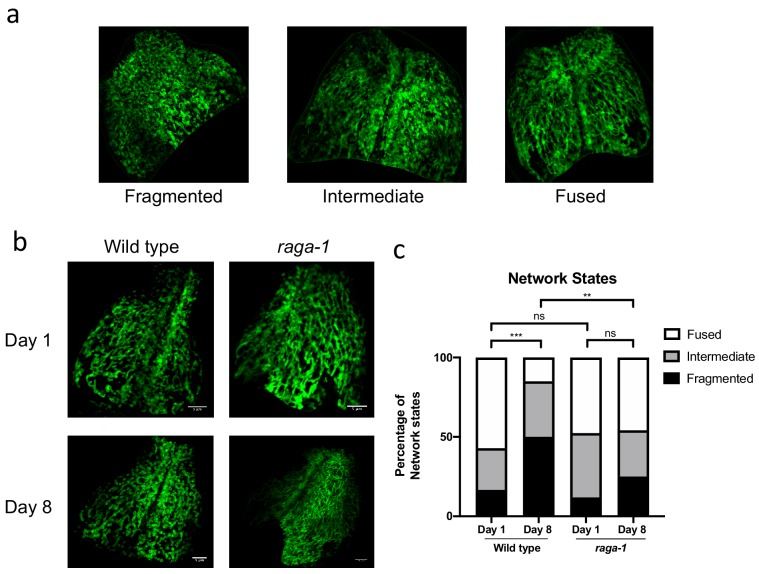
(a) Representative pictures showing that loss of raga-1 preserves muscle mitochondrial content during aging, while neuronal RAGA-1 reverses these effects as seen in the corresponding skeletonized images (Left) post MitoMAPR processing. The mitochondrial skeleton (red) is overlaid on binary images. TOMM-20aa1-49::GFP reporter labels mitochondria in young (day 2) and old (day 11) wild type, raga-1(ok386) mutant and raga-1 neuronal rescue animals (n = 2 independent trials, 16–29 cells were imaged per genotype each time point per replicate). Scale bar represents 25 μm. (b) zoomed insets from (a) (yellow boxes). (c–h) Quantification showing that neuronal raga-1 rescue animals also have decreased mitochondrial coverage (c), greater degree of fragmentation as seen by decreased mitochondrial length (d) and area (e) and increased object number normalized to area (f) compared to raga-1 mutants. Data are represented as mean ±S.D. P value: NS no significance, *<0.05, **<0.01, ***<0.001, between comparisons as indicated by bars. Statistical significance was determined by One-way ANOVA and Welch’s t test. 16–29 cells were quantified per genotype for each time point per replicate (n = 2 independent trials, 25–30 cells were imaged per genotype each time point per replicate). Source data are provided in Figure 4—source data 1.