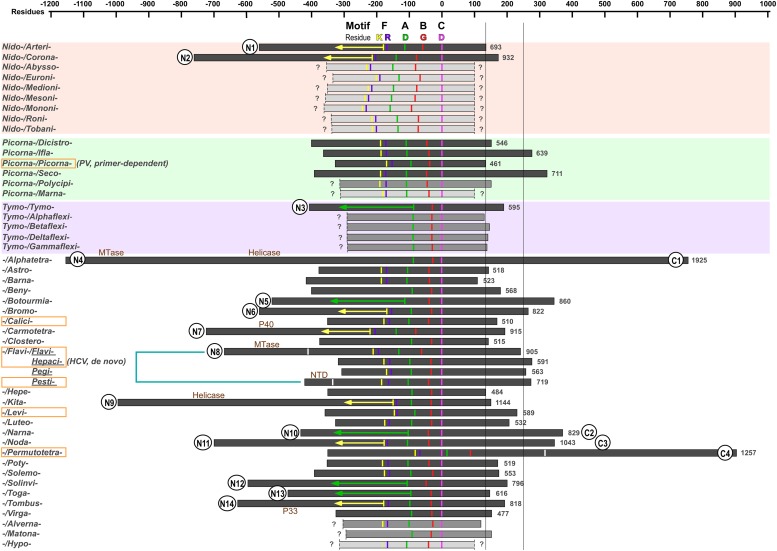
FIGURE 1.
The primary structure comparison of RNA-dependent RNA polymerases (RdRPs) from representative positive-strand RNA viruses. Orders, families, genera, and species assignments based on the ICTV 2018b collection are listed in Table 1. The virus species names are listed in alphabetical order giving the priority to virus order, then to virus family, and then to virus genus. The conserved motif C aspartic acid (magenta, corresponding to PV RdRP D328) is used as the origin in the scale bar. Conserved residues in motifs A, B, and F are also labeled: motif F lysine (corresponding to PV RdRP K159) in yellow; motif F arginine (corresponding to PV RdRP R174) in purple; motif A aspartic acid (corresponding to PV RdRP D233) in green; motif B glycine (corresponding to PV RdRP G289) in red. The orange rectangle indicates that 3D structures are available in that virus family (or virus genus in case of the Flaviviridae). The boundaries of the RdRP catalytic module defined by reported 3D structures are indicated by the white bars for the Flaviviridae and Permutotetraviridae RdRPs. Numbers on the right side of individual RdRP indicate the amino acids numbers for the full-length RdRPs. The question mark (?) indicates undefined boundaries of the RdRP proteins. The yellow and green arrows (150 and 230 residues in length, respectively) are used to estimate the N-terminal boundary of the RdRPs. The two long vertical bars (130 and 250 residues to the origin) indicate the C-terminal boundary of the RdRP catalytic module of the primer-dependent PV 3Dpol and the de novo HCV NS5B and are used to help predict additional functional regions. Wherever available in literature, the name of additional functional regions are labeled. Circles placed at the RdRP termini indicate predicted additional regions. The numbers following the “N” and “C” simply refer to the number of families possibly having additional regions at the RdRP N- and C-termini, respectively.