Figure 2.
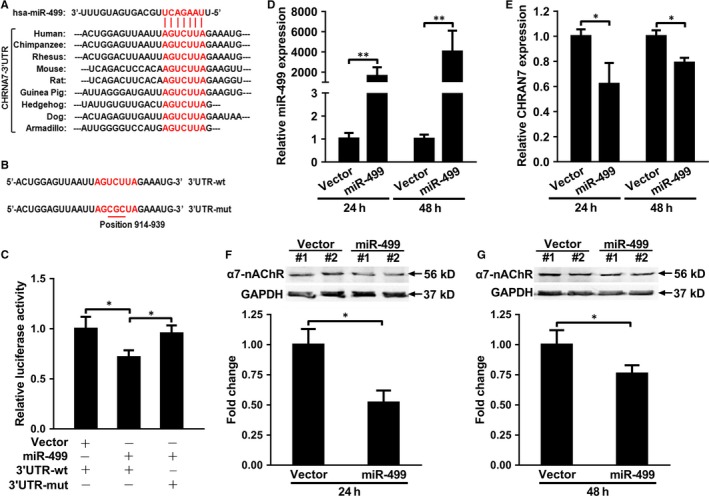
CHRNA7 is a target of hsa‐miR‐499. A, The ‘seed sequence’ of hsa‐miR‐499 is complementary to the target sequence of 3′UTR of CHRNA7 that is conserved amongst species. B, The ‘target sequence’ of human CHRNA7 was mutated from AGUCUUA (wt, top) to AGCGCUA (mut, bottom) with mutated bases underlined. C, miR‐499 significantly decreased the luciferase activity of 3′UTR‐wt compared to Vector, which was abolished by the mutation of the target sequence (3′UTR‐mut). D, miR‐499 expression was significantly higher in hsa‐miR‐499 than vector transfected A549 cells at 24 and 48 h respectively (n = 3 and P < 0.01 for both). E, miR‐499 significantly decreased CHRNA7 expression compared to vector transfection at 24 and 48 h respectively (n = 3 and P < 0.05 for both). F, Representative image of Western Blot showing the effect of transient transfection of Vector versus miR‐499 on the expression of α7‐nAchR expression (top panel) and that of GAPDH (bottom panel), and quantification that miR‐499 significantly decreased the expression of α7‐nAchR than that of Vector (bottom panel) at 24 h (n = 2 and P < 0.05). G, Representative image of Western Blot showing the effect of transient transfection of Vector versus miR‐499 on the expression of α7‐nAchR expression (top panel) and that of GAPDH (bottom panel), and quantification that miR‐499 significantly decreased the expression of α7‐nAchR than that of Vector (bottom panel) at 48 h (n = 2 and P < 0.05). Luciferase activity was normalized to that of Renilla activity. All experiments were in triplicate and repeated at least three times. The expression of miR‐499 and CHRNA7 was normalized to that of U6 RNA and GAPDH respectively in qPCR and the expression of α7‐nAchR was normalized to that of GAPDH in Western blot. Data are presented as ‘Mean ± SD’. The comparison between two groups was performed using paired or non‐paired Student's t test, with * denoting P < 0.05 and ** P < 0.01
