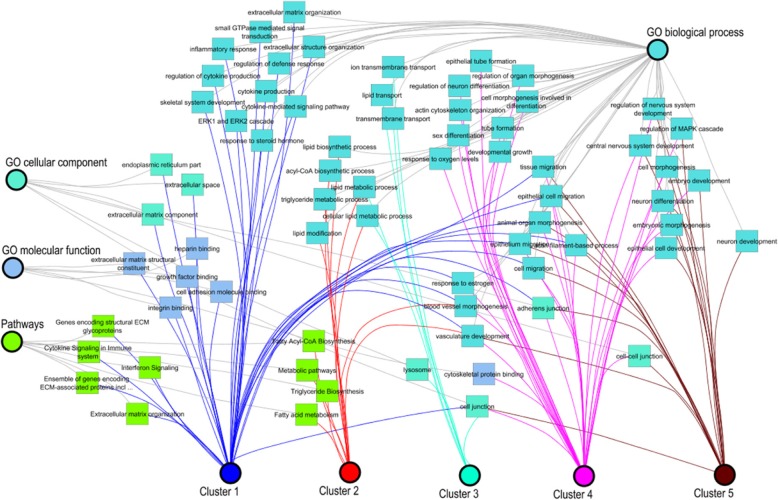
Fig. 4.
Gene Ontology (GO) functional classification network of clusters. All significant differential genes (human Entrez Gene IDs) from clusters were used as input for the ToppCluster. The following databases were used, i.e. “biological process”, “cellular component”, “molecular function” and pathway. Finally, the data were uploaded in Cytoscape 3.6.0 to modify the network. Nodes were colored based on specificity: red nodes specific for different cluster; nodes for the three GO functions and pathway were in different colors