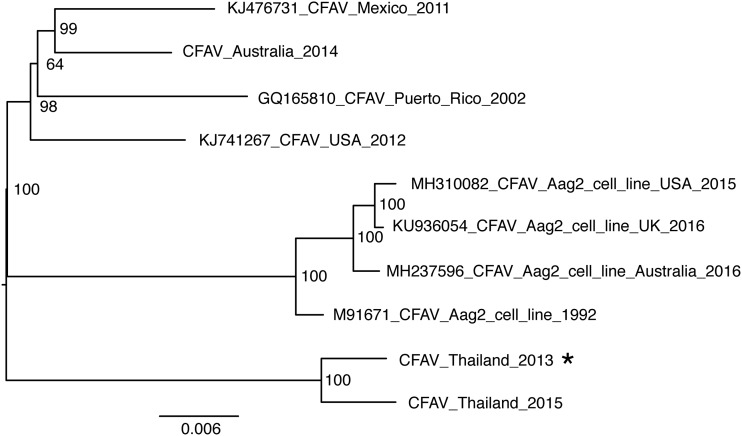
FIG 1.
Phylogenetic relationships among CFAV strains. The tree represents the consensus of 1,000 bootstrap maximum likelihood trees based on the nucleotide alignment of full or nearly full genome sequences of CFAV and a GTR+F+I substitution model. The node support values represent bootstrap proportions. The tree is midpoint rooted, and the scale bar represents the number of nucleotide substitutions per site. The asterisk indicates the newly isolated CFAV strain from this study. GenBank accession numbers are indicated at the beginning of the leaf labels. “Aag2_cell_line” indicates that the virus is derived from the Aag2 cell line, as opposed to live mosquitoes. The country name indicates the geographical origin of mosquitoes from which the virus was isolated or the country where the Aag2 cell line was maintained prior to CFAV sequencing. The year at the end of each name represents the year of CFAV isolation, except for M91671_CFAV_Aag2_cell_line_1992, where 1992 is the year of sequencing since the year of isolation is unknown. Sequences without accession numbers (CFAV_Australia_2014 and CFAV_Thailand_2015) were obtained directly from the authors (12).