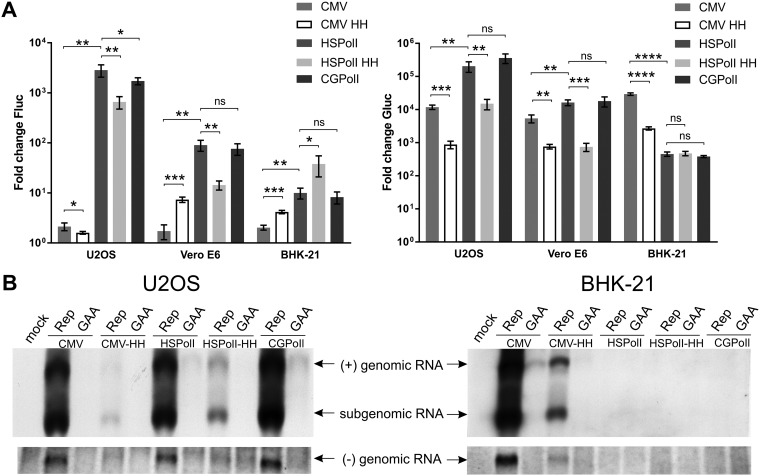
FIG 3.
Comparison of template constructs in mammalian cells. (A) U2OS, Vero E6, and BHK-21 cells were all cotransfected with CMV-P1234 and one of CMV-Fluc-Gluc (CMV), CMV-HH-Fluc-Gluc (CMV HH), HSPolI-Fluc-Gluc (HSPolI), HSPolI-HH-Fluc-Gluc (HSPolI HH), or CGPolI-Fluc-Gluc (CGPolI). Control cells were all cotransfected with CMV-P1234GAA and the same template-expressing plasmids. Cells were lysed at 18 h posttransfection. The Fluc (replication, left) and Gluc (transcription, right) activities generated by the active replicase were normalized to those for the controls. Each column represents an average from three independent experiments; error bars represent standard deviations. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant (Student’s unpaired t test). (B) U2OS and BHK-21 cells were all cotransfected with CMV-P1234 and one of CMV-Fluc-Gluc, CMV-HH-Fluc-Gluc, HSPolI-HH-Fluc-Gluc, HSPolI-Fluc-Gluc, or CGPolI-Fluc-Gluc; control cells were all cotransfected with CMV-P1234GAA and the same template-expressing plasmids or were mock transfected. Samples were collected at 18 h posttransfection. RNA was analyzed by Northern blotting using a probe corresponding to the Fluc reporter gene to detect negative strands (bottom) or a probe complementary to the Gluc reporter gene to detect positive strands (top). “Genomic RNA” designates the full-length template RNA; note that an RNA of the same size is also synthesized by cellular RNA polymerases I and II and is therefore, for some promoters, also detectable in the presence of inactive replicases (GAA). “Subgenomic RNA” designates RNA synthesized by CHIKV replicase using the SG promoter. The experiment was repeated two times with similar results; data from one experiment are shown.