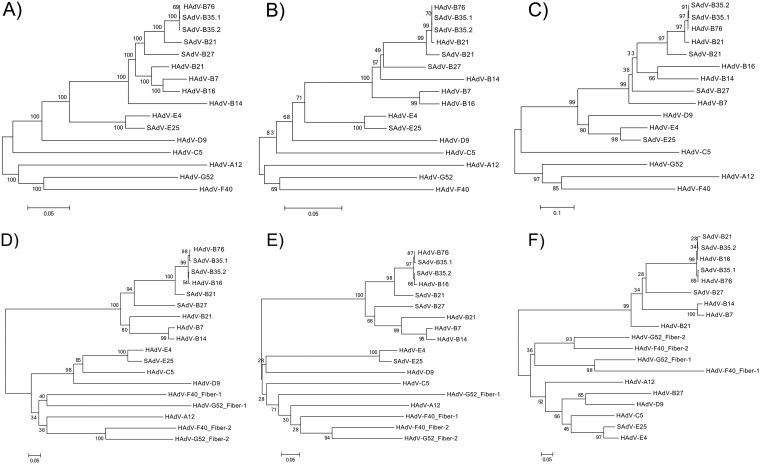
FIG 3.
Phylogenomics analysis of the HAdV-B76 genome and selected genes in the context of the recombinant genome. To provide a clear view of the recombination events, neighbor-joining bootstrap-confirmed phylogenetic trees of the whole-genome (A), penton base (B), hexon (C), and fiber (D) sequences are presented. The fiber gene (D) was further dissected for the detailed analyses of two domains: the shaft, encoded by the proximal sequences (716 bp) (E), and the knob (F), encoded by the distal sequences that include the variable region specifying the hemagglutination inhibition epitope (gamma) and dictating cell tropism (346 bp). The fiber gene appeared recombinant, so domains were extracted using the HAdV-B76 gene sequence and its bootscan profile as a reference, as the shaft and knob lengths varied per HAdV species. These phylogenetic trees were generated after multiple-sequence alignments using the MAFFT software (http://www.ebi.ac.uk/tools/mafft) and default parameters. Bootstrapped, neighbor-joining trees with 1,000 replicates were constructed using MEGA 5.1 software (http://megasoftware.net) with a maximum-composite-likelihood model. Bootstrap values depicted at the nodes indicate the percentages of 1,000 replications producing the clade, with values above 80 considered robust. The scale bar is in units of nucleotide substitutions per site.