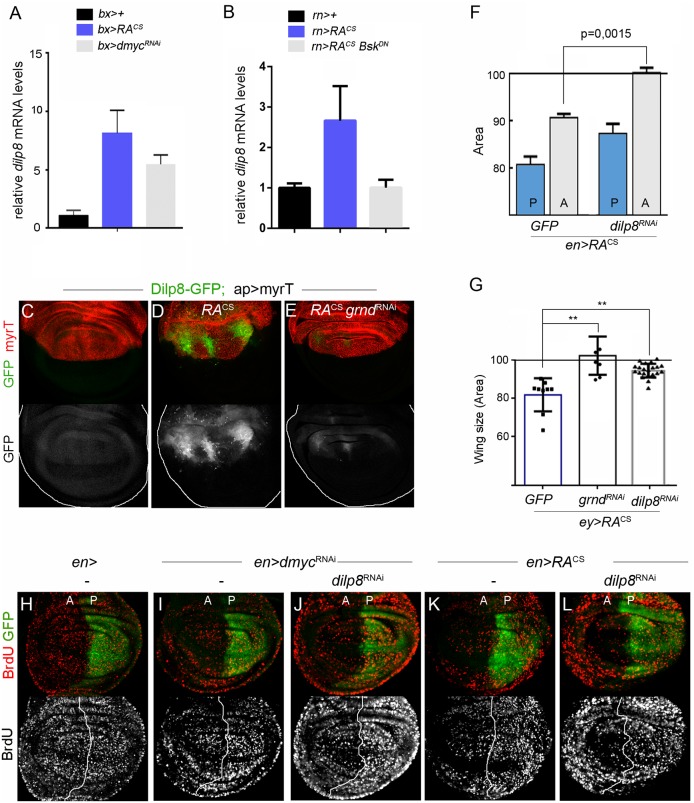
Fig 5. JNK-dependent Dilp8 expression reduces the size of adjacent cell populations.
(A-B) qRT-PCR showing dilp8 transcript levels in wing discs (A) or full larvae (B) expressing the indicated transgenes under the control of bx- or rn-Gal4. Results are expressed as fold induction respect to control wing discs. (C-E) Wing discs carrying a Dilp8-GFP protein trap and expressing the indicated transgenes with ap-Gal4 (expressed in dorsal compartment) were labeled to visualize GFP (green or grey) and myrTomato (red). (F) Histogram plotting normalized area of the P (blue bars) and A (grey bars) compartments of adult wings from individuals expressing RACS alone or in combination with dilp8RNAi using en-Gal4. (G) Histogram plotting normalized wing size from individuals expressing RACS along with the indicated transgenes with ey-Gal4 (drives expression to the eye imaginal disc). Horizontal line shows the size of the normalized control wings (ey-Gal4/UAS-GFP). (H-L) BrdU incorporation assay in larval wing discs from individuals expressing GFP along with the indicated transgenes under the control of en-Gal4. Note the non-autonomous reduction in BrdU incorporation levels caused by dmycRNAi and RACS expression in Dilp8 knock-down wing discs.