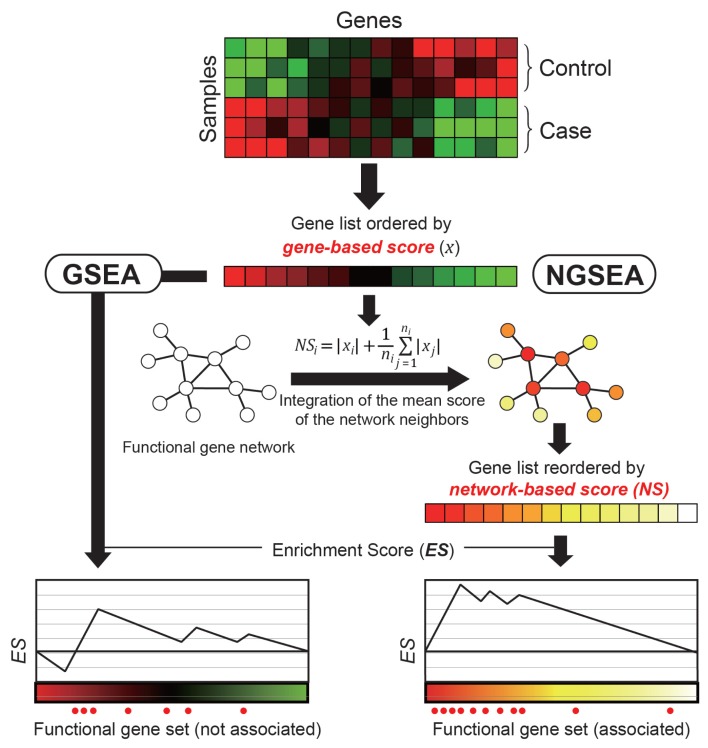
Fig. 1. Overview of NGSEA.
In the original GSEA, genes of a given expression dataset are ordered by gene-based scores (e.g., signal-to-noise ratio [SNR] or log2(Ratio)) based on the gene expression difference between control and case samples. In NGSEA, the genes are ordered by network-based scores, which integrates the gene-based score with the mean of the scores of its neighbors in the functional gene network. This method is based on the observation that functional genes tend to cause expression changes in their network neighbors and therefore are more likely to be correlated with network-based scores.