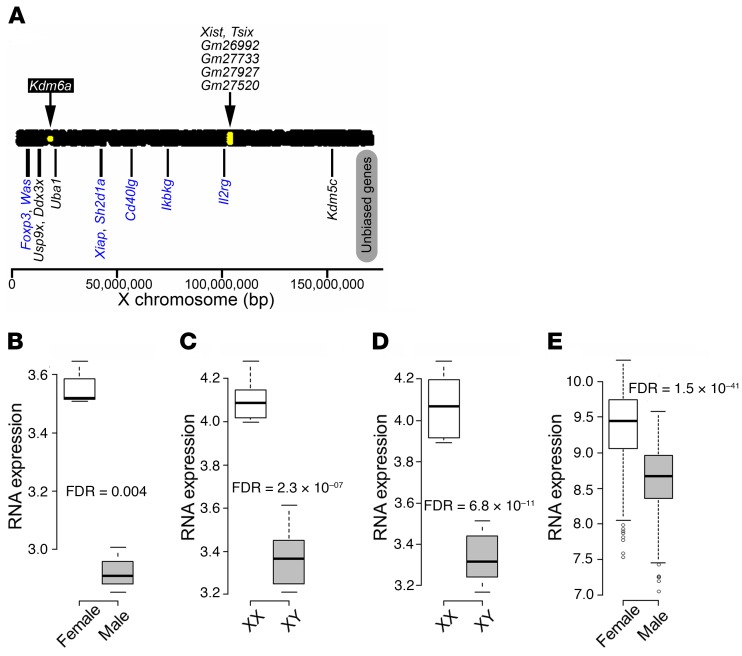
Figure 1. Differentially expressed X chromosome genes in mouse CD4+ T cells.
(A) Using the high-throughput sequencing approach, sexually biased X chromosome genes in CD4+ T cells were determined for C57BL/6J naive CD4+ T cells from spleen (GSE94671; 3 males and 3 females). Yellow dots within the bar representing the X chromosome indicate genes with higher expression in females than in males (threshold for significance was FDR < 0.1). Kdm6a showed increased expression in CD4+ T cells from females as compared with males. Xist and Tsix, which regulate X inactivation, and 4 predicted genes with unknown functions (Gm26992, Gm27733, Gm27927, and Gm27520) also showed increased expression in females. Shown below the bar representing the X chromosome are previously reported X escapees found in other tissues (black text) as well as X genes not thought to escape X inactivation, but involved in immunity (blue text) (58, 59). None had significantly different expression. (B–E) Expression of Kdm6a in (B) naive CD4+ T cells from C57BL/6J spleen (GSE94671; 3 males and 3 females); (C) stimulated CD4+ T cells from C57BL/6J lymph nodes (GSE121292; FCG: 6 XX and 6 XY‾); (D) stimulated CD4+ T cells from SJL lymph nodes (GSE121705; FCG: 6 XX and 5 XY‾); and (E) human naive CD4+ T cells from healthy control blood (GSE56033; 205 males and 294 females). In box-and-whisker plots, thick lines inside the boxes represent the median of the data. The lower and upper ends of boxes show quantiles (25% and 75%), and whiskers show the minimum and maximum values excluding outliers (circles). FDR was calculated using R package edgeR for A–D. For E, 1-way ANOVA was used.