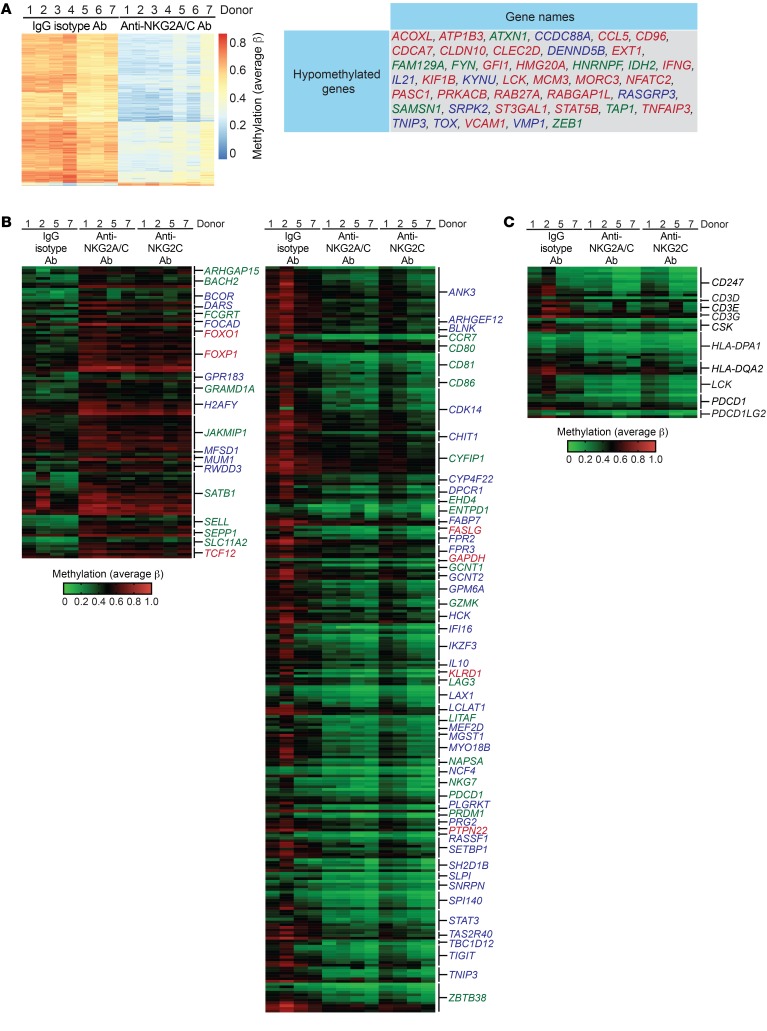
Figure 8. Chronically stimulated adaptive NK cells exhibit a whole-genome DNA methylation profile indicative of exhaustion.
CD3/CD19-depleted PBMCs from HCMV-seropositive donors were cultured for 7 days with 10 ng/ml IL-15 and either IgG2b isotype Ab, anti-NKG2A/C Ab, or anti-NKG2C Ab. The whole-genome DNA methylation profiles of NK cells from these cultures were analyzed using Illumina Infinium MethylationEPIC BeadChips in 2 independent experiments. (A) Heatmap of annotated genomic sites that exhibited consistent differential methylation across NK cells from 7 donors from IgG2b isotype Ab and anti-NKG2A/C Ab cultures and a list of genes within this data set that match genes shown to be differentially expressed or epigenetically remodeled as a result of CD8+ T cell exhaustion. Genes in blue text were shown to be remodeled in exhausted CD8+ T cells as determined by assay for transposase-accessible chromatin with sequencing (ATAC-Seq) (37). Genes in red text were found to be induced in tumor-specific CD8+ T cells as determined by RNA-Seq (34). Genes in green text were identified in both studies. (B) Heatmap of gene-associated promoter regions that exhibited differential methylation between NK cells cultured with IgG2b isotype Ab, anti-NKG2A/C Ab, or anti-NKG2C Ab and were also reported to be differentially regulated in exhausted CD8+ T cells. (C) Heatmap of gene-associated promoter regions matching to the PD-1 signaling pathway as determined by GSEA.