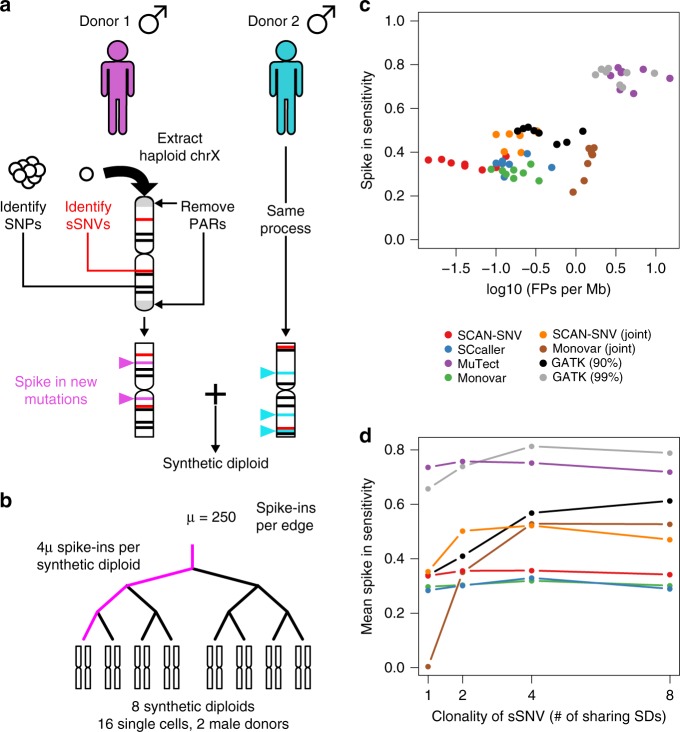
Fig. 5.
Synthetic diploid construction and performance. a Synthetic diploid X chromosomes (chrX) are generated by merging chrX reads from single cells of two male donors. After chrX reads are extracted, pseudoautosomal regions (PARs) are removed and SNPs and sSNVs are identified on the hemizygous regions of chrX. New, randomly placed somatic mutations are spiked into the reads. This process is repeated for a second male donor and the two sets of reads are merged to create a synthetic diploid (SD). b Each SD contains 1000 spike-in mutations with 750 mutations shared by other SDs. For example, 250 spike-ins are shared by all SDs, another 250 are shared by four SDs and so on. c Each point represents genotyper performance on one SD. Only spike-in mutations are used to compute sensitivity. False positive rate is the number of FPs divided by the number of non-PAR megabases on chrX. d Spike-in sensitivity for private spike-ins (clonality = 1), spike-ins shared by two samples (clonality = 2), etc