Fig. 6.
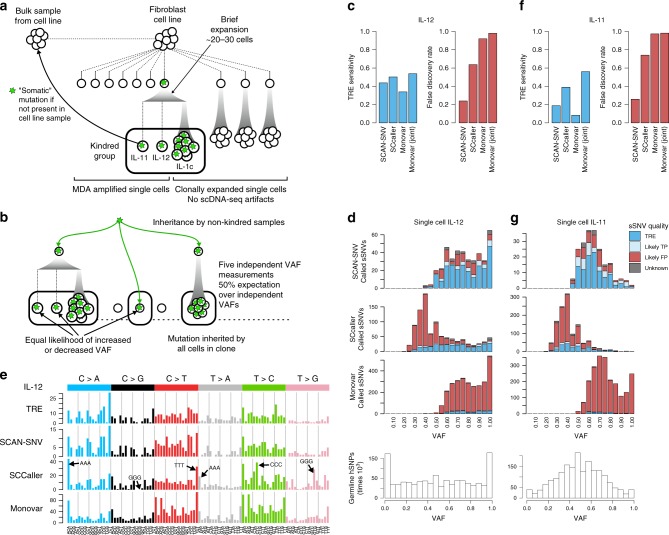
SCAN-SNV performance assessed by a kindred cell system. a Twelve single cell-derived samples from a human fibroblast cell line10. Somatic mutations are defined as mutations not observed in the cell line bulk. A kindred cell system comprising three very closely related samples mimics biological replicates of single cells and enables assessment. True mutations (green stars) are likely to be supported by several kindred samples; scDNA-seq artifacts, however, should be private. b sSNVs in kindred samples with non-kindred support may be subclonal sSNVs. If true, each single cell sample inheriting the subclonal sSNV provides an independent VAF measurement. For true subclonal mutations, the average VAF over many samples should be ~50%. c Single cell genotyper performance on kindred cell IL-12. TRE sensitivity, percent of triple exclusive sites (TREs) recovered; FDR, fraction of total calls classified as likely FPs. d sSNV calls for both genotypers are binned by VAF and classified as either TRE, likely TP or likely FP based on which of the 13 samples contain read support for the mutation. Triple exclusive (TRE) sites are high-quality sites supported by all kindred samples and no other samples. Monovar is run in single-sample mode. (Bottom) hSNP VAFs provide a reference distribution for sSNVs. e Trinucleotide mutation signatures for TREs and genotyper calls in kindred cell IL-12. Monovar is run in single-sample mode. Arrows indicate mutation contexts which create homopolymers. f Same as (c) for kindred cell IL-11. g Same as (d) for kindred cell IL-11. The VAF distribution of both hSNPs and TRE sSNVs is very different from IL-12, suggesting substantial differences in amplification or cell quality