Fig. 7.
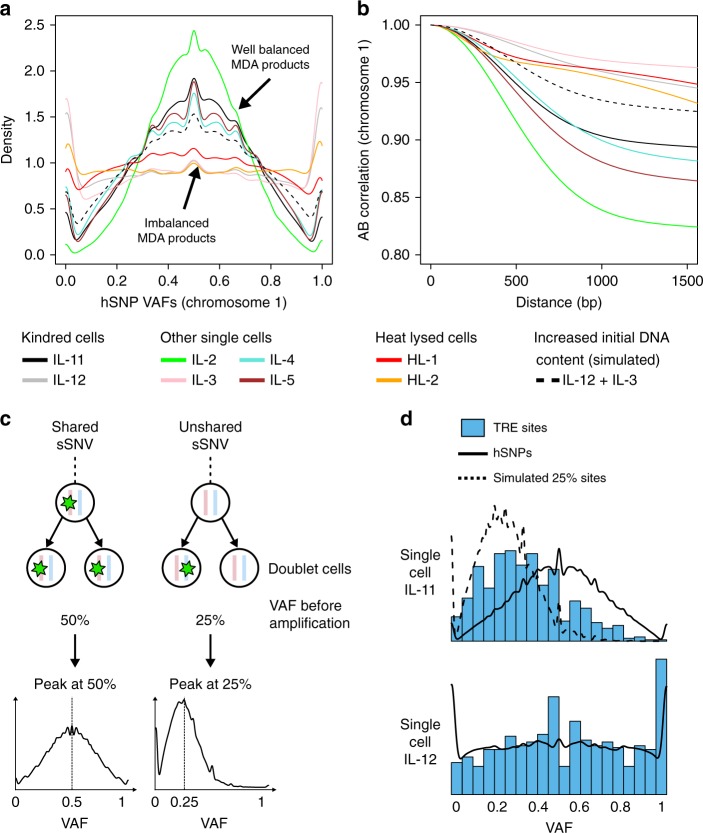
SCAN-SNV recognizes atypical MDA amplification and reveals an accidental doublet. a Chromosome 1 hSNP VAF distributions for eight MDA-amplified single cells fall into one of two classes. Distributions concentrated near half are well-balanced (IL-11-like) whereas flat distribution with peaks near 0 or 1 are imbalanced (IL-12-like). Dashed line: a simulation of a single cell with increased initial DNA content resembles the well-balanced class. b Chromosome 1 AB correlation functions for each MDA-amplified single cell. Well-balanced products are less correlated at short distance than imbalanced products. Dashed line: the increased initial DNA content simulation resembles well-balanced samples. c When two cells are amplified instead of one (an accidental doublet), some somatic mutations will be shared by both cells while others will not. Unshared mutations are supported by only 25% of DNA before amplification. If there are enough unshared somatic SNVs, a doublet sSNV VAF distribution will resemble a mixture of peaks near 25 and 50%. A similar pattern is not expected for a G2/M phase cell. d sSNV VAF distributions are estimated using triple exclusive (TRE) sSNVs (blue). hSNPs show the correct VAF distribution for shared sSNVs and the expectation for unshared sSNVs (dashed line) is obtained by downsampling hSNPs. The peak near VAF = 25% for IL-11 indicates a large number of unshared variants, identifying the sample as an accidental doublet