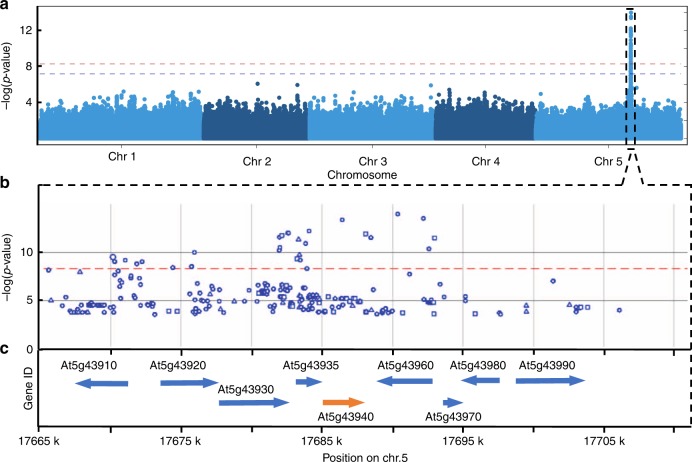
Fig. 2.
Natural GSNOR variants go along with transcription level variation. a Manhattan plots of GSNOR eQTL mapping of 665 Arabidopsis accessions using GWAS analysis. The expression levels of GSNOR gene are depicted in Supplementary Fig. 2f. b, c Regional plots of the significant SNPs b and gene models c. GSNOR is encoded by At5g43940. Circle, triangle, and square denote the SNP to cause intergenic, non-synonymous_coding, and synonymous_coding changes, respectively